Race and genetics: Difference between revisions
→Usage in scientific journals: Added 2021 study of journals |
Unexplained removal |
||
| (129 intermediate revisions by 59 users not shown) | |||
| Line 1: | Line 1: | ||
{{Short description|Relevance of genotype to race classification}} |
{{Short description|Relevance of genotype to race classification}} |
||
{{use dmy dates|date=July 2024}} |
|||
{{cs1 config|name-list-style=vanc|display-authors=4}} |
|||
{{Race}} |
{{Race}} |
||
Researchers have investigated the relationship between '''race and genetics''' as part of efforts to understand how biology may or may not contribute to [[Race (human categorization)|human racial categorization]]. |
|||
Researchers have investigated the relationship between '''race and genetics''' as part of efforts to understand how biology may or may not contribute to [[Race (human categorization)|human racial categorization]]. Today, the consensus among scientists is that race is a [[social construct]], and that using it as a proxy for genetic differences among populations is misleading.<ref name=":8">{{Cite book |url=https://nap.nationalacademies.org/read/26902/chapter/1 |title=Using Population Descriptors in Genetics and Genomics Research: A New Framework for an Evolving Field (Consensus Study Report) |date=2023 |publisher=[[National Academies of Sciences, Engineering, and Medicine]] |doi=10.17226/26902 |pmid=36989389 |isbn=978-0-309-70065-8 |quote=In humans, race is a socially constructed designation, a misleading and harmful surrogate for population genetic differences, and has a long history of being incorrectly identified as the major genetic reason for phenotypic differences between groups.}}</ref><ref name=":4">{{Cite web |date=14 March 2023 |title=Researchers Need to Rethink and Justify How and Why Race, Ethnicity, and Ancestry Labels Are Used in Genetics and Genomics Research, Says New Report |url=https://www.nationalacademies.org/news/2023/03/researchers-need-to-rethink-and-justify-how-and-why-race-ethnicity-and-ancestry-labels-are-used-in-genetics-and-genomics-research-says-new-report |website=National Academies of Sciences, Engineering, and Medicine |quote=Researchers and scientists who utilize genetic and genomic data should rethink and justify how and why they use race, ethnicity, and ancestry labels in their work, says a new National Academies of Sciences, Engineering, and Medicine report. The report says researchers should not use race as a proxy for describing human genetic variation. Race is a social concept, but it is often used in genomics and genetics research as a surrogate for describing human genetic differences, which is misleading, inaccurate, and harmful.}}</ref> |
|||
| ⚫ | Many constructions of race are associated with [[Phenotype|phenotypical]] traits and geographic ancestry, and scholars like [[Carl Linnaeus]] have proposed scientific models for the organization of race since at least the 18th century. Following the discovery of [[Mendelian inheritance|Mendelian genetics]] and the [[Human Genome Project|mapping of the human genome]], questions about the biology of race have often been framed in terms of [[genetics]].<ref name=":3">{{Cite book|last=Goodman|first=Alan H.|url=https://www.worldcat.org/oclc/1121420797|title=Race : are we so different?|date=2020|others=Yolanda T. Moses, Joseph L. Jones|isbn=978-1-119-47247-6|edition=Second|location=Hoboken, NJ|oclc=1121420797}}</ref> A wide range of research methods have been employed to examine patterns of human variation and their relations to ancestry and racial groups, including studies of individual traits,<ref>{{Cite book|last=Jablonski|first=Nina G.|url=https://www.worldcat.org/oclc/64592114|title=Skin : a natural history|date=2006|publisher=University of California Press|isbn=0-520-24281-5|location=Berkeley|oclc=64592114}}</ref> studies of large populations and genetic clusters,<ref>{{Cite journal|last1=Rosenberg|first1=Noah A.|last2=Pritchard|first2=Jonathan K.|last3=Weber|first3=James L.|last4=Cann|first4=Howard M.|last5=Kidd|first5=Kenneth K.|last6=Zhivotovsky|first6=Lev A.|last7=Feldman|first7=Marcus W.|date=2002-12-20|title=Genetic structure of human populations|url=https://pubmed.ncbi.nlm.nih.gov/12493913|journal=Science|volume=298|issue=5602|pages=2381–2385|doi=10.1126/science.1078311|issn=1095-9203|pmid=12493913|bibcode=2002Sci...298.2381R|s2cid=8127224}}</ref> and studies of genetic risk factors for disease.<ref>{{Cite journal|last1=Lorusso|first1=Ludovica|last2=Bacchini|first2=Fabio|date=August 2015|title=A reconsideration of the role of self-identified races in epidemiology and biomedical research|url=https://linkinghub.elsevier.com/retrieve/pii/S1369848615000345|journal=Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences|language=en|volume=52|pages=56–64|doi=10.1016/j.shpsc.2015.02.004|pmid=25791919}}</ref> |
||
| ⚫ | Many constructions of race are associated with [[Phenotype|phenotypical]] traits and geographic ancestry, and scholars like [[Carl Linnaeus]] have proposed scientific models for the organization of race since at least the 18th century. Following the discovery of [[Mendelian inheritance|Mendelian genetics]] and the [[Human Genome Project|mapping of the human genome]], questions about the biology of race have often been framed in terms of [[genetics]].<ref name=":3">{{Cite book|last=Goodman|first=Alan H.|url=https://www.worldcat.org/oclc/1121420797|title=Race : are we so different?|date=2020|others=Yolanda T. Moses, Joseph L. Jones|isbn=978-1-119-47247-6|edition=Second|location=Hoboken, NJ|oclc=1121420797|access-date=2021-04-08|archive-date=2021-05-25|archive-url=https://web.archive.org/web/20210525024950/https://www.worldcat.org/title/race-are-we-so-different/oclc/1121420797|url-status=live |publisher=Wiley Blackwell}}</ref> A wide range of research methods have been employed to examine patterns of human variation and their relations to ancestry and racial groups, including studies of individual traits,<ref>{{Cite book|last=Jablonski|first=Nina G.|url=https://www.worldcat.org/oclc/64592114|title=Skin : a natural history|date=2006|publisher=University of California Press|isbn=0-520-24281-5|location=Berkeley|oclc=64592114|access-date=2021-04-08|archive-date=2021-05-25|archive-url=https://web.archive.org/web/20210525033647/https://www.worldcat.org/title/skin-a-natural-history/oclc/64592114|url-status=live}}</ref> studies of large populations and genetic clusters,<ref name='Rosenberg2002'>{{Cite journal|last1=Rosenberg|first1=Noah A.|author-link1=Noah Rosenberg|last2=Pritchard|first2=Jonathan K.|author-link2=Jonathan Pritchard|last3=Weber|first3=James L.|last4=Cann|first4=Howard M.|last5=Kidd|first5=Kenneth K.|author-link5=Kenneth Kidd|last6=Zhivotovsky|first6=Lev A.|last7=Feldman|first7=Marcus W.|author-link7=Marcus Feldman|date=2002-12-20|title=Genetic structure of human populations|url=https://pubmed.ncbi.nlm.nih.gov/12493913|journal=Science|volume=298|issue=5602|pages=2381–2385|doi=10.1126/science.1078311|issn=1095-9203|pmid=12493913|bibcode=2002Sci...298.2381R|s2cid=8127224|access-date=2021-04-08|archive-date=2021-04-30|archive-url=https://web.archive.org/web/20210430225146/https://pubmed.ncbi.nlm.nih.gov/12493913/|url-status=live}}</ref> and studies of genetic risk factors for disease.<ref name='LorussoBacchini'>{{Cite journal|last1=Lorusso|first1=Ludovica|last2=Bacchini|first2=Fabio|date=August 2015|title=A reconsideration of the role of self-identified races in epidemiology and biomedical research|url=https://linkinghub.elsevier.com/retrieve/pii/S1369848615000345|journal=Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences|language=en|volume=52|pages=56–64|doi=10.1016/j.shpsc.2015.02.004|pmid=25791919|access-date=2021-04-08|archive-date=2021-03-08|archive-url=https://web.archive.org/web/20210308195827/https://linkinghub.elsevier.com/retrieve/pii/S1369848615000345|url-status=live}}</ref> |
||
Research into race and genetics has also been criticized as emerging from, or contributing to, [[scientific racism]]. Some have interpreted genetic studies of traits and populations as evidence to justify social inequalities associated with race,<ref>{{Cite book|last=Saini|first=Angela|url=https://www.worldcat.org/oclc/1091260230|title=Superior : the return of race science|date=2019|isbn=978-0-8070-7691-0|location=Boston|oclc=1091260230}}</ref> despite the fact that patterns of human variation have been shown to be mostly [[Cline (biology)|clinal]], with human genetic code being approximately 99.9% identical between individuals, and with no clear boundaries between groups.<ref>{{Cite book|last=Marks|first=Jonathan|url=https://www.worldcat.org/oclc/961801723|title=Is science racist?|date=2017|isbn=978-0-7456-8921-0|location=Malden, MA|oclc=961801723}}</ref><ref name=":3" /> |
|||
Research into race and genetics has also been criticized as emerging from, or contributing to, [[scientific racism]]. Genetic studies of traits and populations have been used to justify [[Racial inequality|social inequalities associated with race]],<ref>{{Cite book|last=Saini|first=Angela|url=https://www.worldcat.org/oclc/1091260230|title=Superior : the return of race science|date=2019|isbn=978-0-8070-7691-0|location=Boston|oclc=1091260230|access-date=2021-04-08|archive-date=2021-08-08|archive-url=https://web.archive.org/web/20210808083306/https://www.worldcat.org/title/superior-the-return-of-race-science/oclc/1091260230|url-status=live |publisher=Beacon Press}}</ref> despite the fact that patterns of human variation have been shown to be mostly [[Cline (biology)|clinal]],<ref>{{Cite book|last=Marks|first=Jonathan|url=https://www.worldcat.org/oclc/961801723|title=Is science racist?|date=2017|isbn=978-0-7456-8921-0|location=Malden, MA|oclc=961801723|access-date=2021-04-08|archive-date=2021-05-02|archive-url=https://web.archive.org/web/20210502185326/https://www.worldcat.org/title/is-science-racist/oclc/961801723|url-status=live |publisher=Polity}}</ref> with human genetic code being approximately 99.6%-99.9% identical between individuals and without clear boundaries between groups.<ref name='Tishkoff&Kidd2004'>{{cite journal |last1=Tishkoff |first1=Sarah A |last2=Kidd |first2=Kenneth K |author1-link=Sarah Tishkoff |author2-link=Kenneth Kidd |title=Implications of biogeography of human populations for 'race' and medicine |journal=Nature Genetics |date=26 October 2004 |volume=36 |issue=11 |doi=10.1038/ng1438 |pmid=15507999 |url=https://www.nature.com/articles/ng1438 |access-date=26 June 2024 |series=Supplemental |pages=S21-7 |publisher=[[Nature Portfolio]] |s2cid=1500915}}</ref> |
|||
There is ongoing scientific debate regarding the definition and meaning of race in genetic and biomedical research. Some researchers argue that race can act as a "proxy" for genetic ancestry because individuals of the same racial category may share a common ancestry, but others advocate for distinguishing between biology and the social, political, cultural, and economic factors that contribute to race as most commonly understood.<ref>{{Cite journal|last1=Bamshad|first1=Michael|last2=Wooding|first2=Stephen|last3=Salisbury|first3=Benjamin A.|last4=Stephens|first4=J. Claiborne|date=August 2004|title=Deconstructing the relationship between genetics and race|url=http://www.nature.com/articles/nrg1401|journal=Nature Reviews Genetics|language=en|volume=5|issue=8|pages=598–609|doi=10.1038/nrg1401|pmid=15266342|s2cid=12378279|issn=1471-0056}}</ref> |
|||
Some researchers have argued that race can act as a proxy for genetic ancestry because individuals of the same racial category may share a common ancestry, but this view has fallen increasingly out of favor among experts.<ref name=":4" /><ref>{{Cite journal |last=Kaiser |first=Jocelyn |date=11 March 2023 |title=Geneticists should rethink how they use race and ethnicity, panel urges |url=https://www.science.org/content/article/geneticists-should-rethink-how-they-use-race-and-ethnicity-panel-urges |journal=Science}}</ref> The mainstream view is that it is necessary to distinguish between biology and the social, political, cultural, and economic factors that contribute to conceptions of race.<ref name='AABAstatement'>{{cite press release |title=AABA (AAPA) Statement on Race & Racism |last1=Ackermann |first1=Rebecca |last2=Athreya |first2=Sheela |last3=Bolnick |first3=Deborah |last4=Fuentes |first4=Agustín |author-link4=Agustín Fuentes |last5=Lasisi |first5=Tina |last6=Lee |first6=Sang-Hee |last7=McLean |first7=Shay-Akil |last8=Nelson |first8=Robin |url=https://physanth.org/about/position-statements/aapa-statement-race-and-racism-2019/ |year=2019 |access-date=2024-06-26}}</ref><ref>{{Cite journal|last1=Bamshad|first1=Michael|last2=Wooding|first2=Stephen|last3=Salisbury|first3=Benjamin A.|last4=Stephens|first4=J. Claiborne|date=August 2004|title=Deconstructing the relationship between genetics and race|url=http://www.nature.com/articles/nrg1401|journal=Nature Reviews Genetics|language=en|volume=5|issue=8|pages=598–609|doi=10.1038/nrg1401|pmid=15266342|s2cid=12378279|issn=1471-0056|access-date=2021-04-08|archive-date=2021-06-10|archive-url=https://web.archive.org/web/20210610120335/https://www.nature.com/articles/nrg1401|url-status=live}}</ref> |
|||
Phenotype may have a tangential connection to DNA, but it is still only a rough proxy that would omit various other genetic information.<ref name=":4" /><ref>{{Cite web |first=Megan |last=Gannon |title=Race Is a Social Construct, Scientists Argue |url=https://www.scientificamerican.com/article/race-is-a-social-construct-scientists-argue/ |access-date=2024-03-12 |website=Scientific American |language=en}}</ref><ref>{{Cite web |website=ScienceNews |first=Tina |last=Hesman Saey|date=2023-03-14 |title=Why experts recommend ditching racial labels in genetic studies |url=https://www.sciencenews.org/article/race-label-genetic-stigma-diversity |access-date=2024-03-12 |language=en-US}}</ref> Today, in a somewhat similar way that "gender" is differentiated from the more clear "biological sex", scientists state that potentially "race" / phenotype can be differentiated from the more clear "ancestry".<ref>{{Cite web |first=Vivian |last=Chou |date=2017-04-18 |title=How Science and Genetics are Reshaping the Race Debate of the 21st Century |url=https://sitn.hms.harvard.edu/flash/2017/science-genetics-reshaping-race-debate-21st-century/ |access-date=2024-03-12 |website=Science in the News |language=en-US}}</ref> However, this system has also still come under scrutiny as it may fall into the same problems – which would be large, vague groupings with little genetic value.<ref>{{Cite web |last=Lewis |first=Anna C. F. |date=2022-05-02 |title=Substituting genetic ancestry for race in research? Not so fast |url=https://www.statnews.com/2022/05/02/substituting-genetic-ancestry-for-race-in-research-not-so-fast/ |access-date=2024-03-12 |website=STAT |language=en-US}}</ref> |
|||
==Overview== |
==Overview== |
||
===The concept of race=== |
===The concept of race=== |
||
{{See also|Race (human categorization)}} |
{{See also|Race (human categorization)}} |
||
The concept of "race" as a classification system of humans based on visible physical characteristics emerged over the last five centuries, influenced by European colonialism.<ref |
The concept of "race" as a classification system of humans based on visible physical characteristics emerged over the last five centuries, influenced by European colonialism.<ref name='AABAstatement'/><ref>{{cite book |last1=Kennedy |first1=Rebecca F.|author-link=Rebecca Futo Kennedy |last2=Roy |first2=C. Sydnor |last3=Goldman |first3=Max L. |date=2013 |title=Race and Ethnicity in the Classical World |location=Indianapolis, Indiana |publisher=Hackett Publishing Company, Inc. |page=xiii |isbn=978-1603849944}}</ref> However, there is widespread evidence of what would be described in modern terms as ''racial consciousness'' throughout the entirety of [[recorded history]]. For example, in [[Ancient Egypt]] there were [[Book of Gates|four broad racial divisions]] of human beings: Egyptians, Asiatics, Libyans, and Nubians.<ref>{{Cite web|title=Race in Ancient Egypt|url=https://www.ucl.ac.uk/museums-static/digitalegypt/social/race.html|access-date=2021-07-31|website=www.ucl.ac.uk|archive-date=2021-07-31|archive-url=https://web.archive.org/web/20210731060151/https://www.ucl.ac.uk/museums-static/digitalegypt/social/race.html|url-status=live}}</ref> There was also [[Aristotle]] of [[Ancient Greece]], who once wrote: "The peoples of Asia... lack spirit, so that they are in continuous subjection and slavery."<ref>{{Cite web|title=Aristotle, Politics, Book 7, section 1327b|url=http://www.perseus.tufts.edu/hopper/text?doc=Perseus:abo:tlg,0086,035:7:1327b|access-date=2021-07-31|website=www.perseus.tufts.edu|archive-date=2021-07-31|archive-url=https://web.archive.org/web/20210731060150/http://www.perseus.tufts.edu/hopper/text?doc=Perseus:abo:tlg,0086,035:7:1327b|url-status=live}}</ref> The concept has [[Historical race concepts|manifested in different forms]] based on social conditions of a particular group, often used to justify unequal treatment. Early influential attempts to classify humans into discrete races include 4 races in Carl Linnaeus's ''Systema Naturae'' (''Homo europaeus'', ''asiaticus'', ''americanus'', and ''afer'')<ref>{{cite book |last1=Slotkin |first1=James S. |date=1965 |title=Readings in Early Anthropology |location=London |publisher=Routledge |isbn=9780203715215}}</ref><ref>{{cite book| last1=Linnaeus |first1=C. |date=1758 |title=Systema naturae |location=Stockholm |publisher=Laurentii Salvii|page=532}}</ref> and 5 races in Johann Friedrich Blumenbach's ''On the Natural Variety of Mankind''.<ref>{{cite book |last1=Blumenbach |first1=J.F. |last2=Bendyshe |first2=T.T. |date=1795 |title=On the natural variety of mankind}}</ref> Notably, over the next centuries, scholars argued for anywhere from 3 to more than 60 race categories.<ref>{{cite book| last1=Darwin |first1=Charles| date=1871| title=The Descent of Man, and Selection in Relation to Sex}}</ref> Race concepts have changed within a society over time; for example, in the United States social and legal designations of "White" have been inconsistently applied to Native Americans, Arab Americans, and Asian Americans, among other groups (''See main article: [[Definitions of whiteness in the United States]]''). Race categories also vary worldwide; for example, the same person might be perceived as belonging to a different category in the United States versus Brazil.<ref>{{cite book| last1=Daniel |first1=G.R. |date=2006 |title=Race and multiraciality in Brazil and the United States: converging paths? |publisher=Penn State Press}}</ref> Because of the arbitrariness inherent in the concept of race, it is difficult to relate it to biology in a straightforward way. |
||
===Race and human genetic variation=== |
===Race and human genetic variation=== |
||
There is broad consensus across the biological and social sciences that race is a social construct, not an accurate representation of human genetic variation.<ref name="Yudell2016">{{cite journal |last1=Yudell |first1=Michael |last2=Roberts |first2=Dorothy |last3=DeSalle |first3=Rob |last4=Tishkoff |first4=Sarah |author4-link=Sarah Tishkoff |title=Taking race out of human genetics |journal=[[Science (journal)|Science]] |date=2016 |volume=351 |issue=6273 |pages=564–565 |doi=10.1126/science.aac4951 |url=https://www.science.org/doi/abs/10.1126/science.aac4951 |access-date=27 June 2024 |publisher=[[American Association for the Advancement of Science]] |issn=0036-8075 |pmid=26912690 |oclc=6005630581 |s2cid=206639306 |bibcode=2016Sci...351..564Y |archive-url=https://web.archive.org/web/20210913114037/https://www.science.org/doi/abs/10.1126/science.aac4951 |archive-date=13 September 2021}}</ref><ref name='Tishkoff&Kidd2004'/> As more progress has been made on sequencing the human genome, it has been found that any two humans will share an average of 99.35% of their DNA based on the approximately 3.1 billion haploid base pairs.<ref name="Auton2015">{{cite journal |last1=Auton |first1=Adam |last2=Abecasis |first2=Gonçalo R. |last3=Altshuler |first3=David M. |last4=Durbin |first4=Richard M. |last5=Bentley |first5=David R. |last6=Chakravarti |first6=Aravinda |last7=Clark |first7=Andrew G. |last8=Donnelly |first8=Peter |last9=Eichler |first9=Evan E. |last10=Flicek |first10=Paul |last11=Gabriel |first11=Stacey B. |last12=Gibbs |first12=Richard A. |last13=Green |first13=Eric D. |last14=Hurles |first14=Matthew E. |last15=Knoppers |first15=Bartha M. |last16=Korbel |first16=Jan O. |last17=Lander |first17=Eric S. |last18=Lee |first18=Charles |last19=Lehrach |first19=Hans |last20=Mardis |first20=Elaine R. |last21=Marth |first21=Gabor T. |last22=McVean |first22=Gil A. |last23=Nickerson |first23=Deborah A. |last24=Schmidt |first24=Jeanette P. |last25=Sherry |first25=Stephen T. |last26=Wang |first26=Jun |last27=Wilson |first27=Richard K. |last28=Boerwinkle |first28=Eric |last29=Doddapaneni |first29=Harsha |last30=Han |first30=Yi |last31=Korchina |first31=Viktoriya |last32=Kovar |first32=Christie |last33=Lee |first33=Sandra |last34=Muzny |first34=Donna |last35=Reid |first35=Jeffrey G. |last36=Zhu |first36=Yiming |last37=Chang |first37=Yuqi |last38=Feng |first38=Qiang |last39=Feng |first39=Xiaodong |last40=Guo |first40=Xiaosen |last41=Jian |first41=Min |last42=Jiang |first42=Hui |last43=Jin |first43=Xin |last44=Lan |first44=Tianming |last45=Li |first45=Guoqing |last46=Li |first46=Jingxiang |last47=Liu |first47=Shengmao |last48=Liu |first48=Xiao |last49=Lu |first49=Yao |last50=Ma |first50=Xuedi |last51=Tang |first51=Meifang |last52=Wang |first52=Bo |last53=Wang |first53=Guangbiao |last54=Wu |first54=Honglong |last55=Wu |first55=Renhua |last56=Xu |first56=Xun |last57=Yin |first57=Ye |last58=Zhang |first58=Dandan |last59=Zhang |first59=Wenwei |last60=Zhao |first60=Jiao |last61=Zhao |first61=Meiru |last62=Zheng |first62=Xiaole |last63=Gupta |first63=Namrata |last64=Gharani |first64=Neda |last65=Toji |first65=Lorraine H. |last66=Gerry |first66=Norman P. |last67=Resch |first67=Alissa M. |last68=Barker |first68=Jonathan |last69=Clarke |first69=Laura |last70=Gil |first70=Laurent |last71=Hunt |first71=Sarah E. |last72=Kelman |first72=Gavin |last73=Kulesha |first73=Eugene |last74=Leinonen |first74=Rasko |last75=McLaren |first75=William M. |last76=Radhakrishnan |first76=Rajesh |last77=Roa |first77=Asier |last78=Smirnov |first78=Dmitriy |last79=Smith |first79=Richard E. |last80=Streeter |first80=Ian |last81=Thormann |first81=Anja |last82=Toneva |first82=Iliana |last83=Vaughan |first83=Brendan |last84=Zheng-Bradley |first84=Xianggun |last85=Grocock |first85=Russell |last86=Humphray |first86=Sean |last87=James |first87=Terena |last88=Kingsbury |first88=Zoya |last89=Sudbrak |first89=Ralf |last90=Albrecht |first90=Marcus W. |last91=Amstislavskiy |first91=Vyacheslav S. |last92=Borodina |first92=Tatiana A. |last93=Lienhard |first93=Matthias |last94=Mertes |first94=Florian |last95=Sultan |first95=Marc |last96=Timmermann |first96=Bernd |last97=Yaspo |first97=Marie-Laure |last98=Fulton |first98=Lucinda |last99=Fulton |first99=Robert |last100=Ananiev |first100=Victor |last101=Belaia |first101=Zinaida |last102=Beloslyudtsev |first102=Dimitriy |last103=Bouk |first103=Nathan |last104=Chen |first104=Chao |last105=Church |first105=Deeanna |last106=Cohen |first106=Robert |last107=Cook |first107=Charles |last108=Garner |first108=John |last109=Hefferon |first109=Timothy |last110=Kimelman |first110=Mikhail |last111=Liu |first111=Chunlei |last112=Lopez |first112=John |last113=Meric |first113=Peter |last114=O'Sullivan |first114=Chris |last115=Ostapchuk |first115=Yuri |last116=Phan |first116=Lon |last117=Ponomarov |first117=Sergiy |last118=Schneider |first118=Valerie |last119=Shekhtman |first119=Eugene |last120=Sirotkin |first120=Karl |last121=Slotta |first121=Douglas |last122=Zhang |first122=Hua |last123=Balasubramaniam |first123=Senduran |last124=Burton |first124=John |last125=Danecek |first125=Petr |last126=Keane |first126=Thomas M. |last127=Kolb-Kokocinski |first127=Anja |last128=McCarthy |first128=Shane |last129=Stalker |first129=James |last130=Quail |first130=Michael |last131=Gollub |first131=Jeremy |last132=Webster |first132=Teresa |last133=Wong |first133=Brant |last134=Zhan |first134=Yiping |last135=Campbell |first135=Christopher L. |last136=Kong |first136=Yu |last137=Marcketta |first137=Anthony |last138=Yu |first138=Fuli |last139=Antunes |first139=Lilian |last140=Bainbridge |first140=Matthew |last141=Sabo |first141=Aniko |last142=Huang |first142=Zhuoyi |last143=Coin |first143=Lachlan J. M. |last144=Fang |first144=Lin |last145=Li |first145=Qibin |last146=Li |first146=Zhenyu |last147=Lin |first147=Haoxiang |last148=Liu |first148=Binghang |last149=Luo |first149=Ruibang |last150=Shao |first150=Haojing |last151=Xie |first151=Yinlong |last152=Ye |first152=Chen |last153=Yu |first153=Chang |last154=Zhang |first154=Fan |last155=Zheng |first155=Hancheng |last156=Zhu |first156=Hongmei |last157=Alkan |first157=Can |last158=Dal |first158=Elif |last159=Kahveci |first159=Fatma |last160=Garrison |first160=Erik P. |last161=Kural |first161=Deniz |last162=Lee |first162=Wan-Ping |last163=Fung Leong |first163=Wen |last164=Stomberg |first164=Michael |last165=Ward |first165=Alistair N. |last166=Wu |first166=Jiantao |last167=Zhang |first167=Mengyao |last168=Daly |first168=Mark J. |last169=DePristo |first169=Mark A. |last170=Handsaker |first170=Robert E. |last171=Banks |first171=Eric |last172=Bhatia |first172=Gaurav |last173=del Angel |first173=Guillermo |last174=Genovese |first174=Giulio |last175=Li |first175=Heng |last176=Kashin |first176=Seva |last177=McCarroll |first177=Steven A. |last178=Nemesh |first178=James C. |last179=Poplin |first179=Ryan E. |last180=Yoon |first180=Seungtai C. |last181=Lihm |first181=Jayon |last182=Makarov |first182=Vladimir |last183=Gottipati |first183=Srikanth |last184=Keinan |first184=Alon |last185=Rodriguez-Flores |first185=Juan L. |last186=Rausch |first186=Tobias |last187=Fritz |first187=Markus H. |last188=Stütz |first188=Adrian M. |last189=Beal |first189=Kathryn |last190=Datta |first190=Avik |last191=Herrero |first191=Javier |last192=Ritchie |first192=Graham R. S. |last193=Zerbino |first193=Daniel |last194=Sabeti |first194=Pardis C. |last195=Shlyakhter |first195=Ilya |last196=Schaffner |first196=Stephen F. |last197=Vitti |first197=Joseph |last198=Cooper |first198=David N. |last199=Ball |first199=Edward V. |last200=Stenson |first200=Peter D. |last201=Barnes |first201=Bret |last202=Bauer |first202=Markus |last203=Keira Cheetham |first203=R. |last204=Cox |first204=Anthony |last205=Eberle |first205=Michael |last206=Kahn |first206=Scott |last207=Murray |first207=Lisa |last208=Pedan |first208=John |last209=Shaw |first209=Richard |last210=Kenny |first210=Eimear E. |last211=Batzer |first211=Mark A. |last212=Konkel |first212=Miriam K. |last213=Walker |first213=Jerilyn A. |last214=MacArthur |first214=Daniel G. |last215=Lek |first215=Monkol |last216=Herwig |first216=Ralf |last217=Ding |first217=Li |last218=Koboldt |first218=Daniel C. |last219=Larson |first219=David |last220=Ye |first220=Kai |last221=Gravel |first221=Simon |author2-link=Gonçalo Abecasis |author3-link=David Altshuler (physician) |author4-link=Richard M. Durbin |author6-link=Aravinda Chakravarti |author7-link=Andrew G. Clark |author8-link=Peter Donnelly |author9-link=Evan E. Eichler |author11-link=Stacey Gabriel |author12-link=Richard Gibbs (biologist) |author13-link=Eric D. Green |author14-link=Matthew Hurles |author15-link=Bartha Knoppers |author16-link=Jan O. Korbel |author17-link=Eric Lander |author18-link=Charles Lee (scientist) |author20-link=Elaine Mardis |author22-link=Gil McVean |author23-link=Deborah Nickerson |author26-link=Wang Jun (scientist) |author27-link=Richard K. Wilson |title=A global reference for human genetic variation |journal=Nature |date=October 2015 |volume=526 |issue=7571 |pages=68–74 |doi=10.1038/nature15393 |pmid=26432245 |publisher=[[Nature Portfolio]] |issn=1476-4687 |pmc=4750478 |bibcode=2015Natur.526...68T |oclc=8521848829 |id=Auton2015 |display-authors=1}}</ref><ref name="Piovesan2019">{{cite journal |last1=Piovesan |first1=Allison |last2=Chiara Pelleri |first2=Maria |last3=Antonaros |first3=Francesca |last4=Strippoli |first4=Pierluigi |last5=Caracausi |first5=Maria |last6=Vitale |first6=Lorenza |title=On the length, weight and GC content of the human genome |journal=BMC Research Notes |date=February 2019 |volume=12 |issue=1 |page=106 |doi=10.1186/s13104-019-4137-z |pmid=30813969 |publisher=[[BioMed Central]] |doi-access=free |issn=1756-0500 |pmc=6391780 |oclc=8016439744}}</ref> However, this number should be understood as an average, any two specific individuals can have their genomes differ by more or less than 0.65%. Additionally, this average is an estimate, subject to change as additional sequences are discovered and populations sampled. In 2010, the genome of [[Craig Venter]] was found to differ by an estimated 1.59% from a [[reference genome]] created by the [[National Center for Biotechnology Information]].<ref name="Pang2010">{{cite journal |last1=Pang |first1=Andy W. |last2=MacDonald |first2=Jeffrey R. |last3=Pinto |first3=Dalila |last4=Wei |first4=John |last5=Rafiq |first5=Muhammad A. |last6=Conrad |first6=Donald F. |last7=Park |first7=Hansoo |last8=Hurles |first8=Matthew E. |author8-link=Matthew Hurles |last9=Lee |first9=Charles |author9-link=Charles Lee (scientist) |last10=Venter |first10=J. Craig |author10-link=Craig Venter |last11=Kirkness |first11=Ewan F. |last12=Levy |first12=Samuel |last13=Feuk |first13=Lars |last14=Scherer |first14=Stephen W. |author14-link=Stephen W. Scherer |title=Towards a comprehensive structural variation map of an individual human genome |journal=[[Genome Biology]] |date=May 2010 |volume=11 |issue=5 |page=R52 |doi=10.1186/gb-2010-11-5-r52 |pmid=20482838 |publisher=[[BioMed Central]] |doi-access=free |issn=1474-760X |pmc=2898065 |oclc=5660396679 |id=Pang2010}}</ref> <!--to do: add discussion of human genetic diversity in comparison to chimpanzees--> |
|||
There is broad consensus across the biological and social sciences that race is a social construct, not an accurate representation of human genetic variation.<ref name="AAPA statement on race and racism">{{cite journal|last1=Fuentes|first1=A|last2=Ackermann|first2=RR|last3=Athreya|first3=S|last4=Bolnik|first4=D|last5=Lasisi|first5=T|last6=Lee|first6=S|last7=McLean|first7=S|last8=Nelson|first8=Robin|date=2019|title=AAPA statement on race and racism|url=https://onlinelibrary.wiley.com/doi/full/10.1002/ajpa.23882|journal=American Journal of Physical Anthropology|volume=169|issue=3|pages=400–402|doi=10.1002/ajpa.23882|pmid=31199004|s2cid=189815619|access-date=21 June 2021}}</ref><ref>{{cite journal |last1=Yudell |first1=M |last2=Roberts |first2=D |last3=DeSalle |first3=R |last4=Tishkoff |first4=S |date=2016 |title=Science and society: Taking race out of human genetics |url=https://pubmed.ncbi.nlm.nih.gov/26912690/ |journal=Science |volume=351 |issue=6273 |pages=564–565 |doi=10.1126/science.aac4951|pmid=26912690 |s2cid=206639306 }}</ref><ref name="Implications of biogeography of hum">{{cite journal |last1=Tishkoff |first1=SA |last2=Kidd |first2=KK |date=2004 |title=Implications of biogeography of human populations for 'race' and medicine |journal=Nature Genetics |volume=36 |issue=11 Suppl |pages=s21–s27|doi=10.1038/ng1438 |pmid=15507999 |s2cid=1500915 |doi-access=free }}</ref> Humans are remarkably genetically similar, sharing approximately 99.9% of their genetic code with one another. We nonetheless see wide individual variation in phenotype, which arises from both genetic differences and complex gene-environment interactions. The vast majority of this genetic variation occurs ''within'' groups; very little genetic variation differentiates ''between'' groups.<ref>{{cite journal |last1=Rosenberg |first1=N.A. |date=2002 |title=Genetic structure of human populations |url=https://pubmed.ncbi.nlm.nih.gov/12493913/ |journal=Science |volume=298 |issue=5602 |pages=2381–2385 |doi=10.1126/science.1078311|pmid=12493913 |bibcode=2002Sci...298.2381R |s2cid=8127224 }}</ref> Crucially, the between-group genetic differences that do exist do not map onto socially recognized categories of race. Furthermore, although human populations show some genetic clustering across geographic space, human genetic variation is "[[Cline (biology)|clinal]]", or continuous.<ref name="AAPA statement on race and racism"/><ref name="Implications of biogeography of hum"/> This, in addition to the fact that different traits vary on different clines, makes it impossible to draw discrete genetic boundaries around human groups. Finally, insights from ancient DNA are revealing that no human population is "pure" – all populations represent a long history of migration and mixing.<ref>{{cite book| last1=Reich |first1=David |date=2018 |title=Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past |publisher=Oxford University Press}}</ref> For example, the genetic makeup of European populations was massively transformed by waves of migrations of farmers from the Near East between 8,500-5,000 years ago and Yamnaya pastoralists from the Eurasian steppe beginning around 4,500 years ago.<ref>{{cite journal |last1=Haak |first1=W |last2=Lazaridis |first2=I|date=2015 |title=Massive migration from the steppe was a source for Indo-European languages in Europe |journal=Nature |volume=522 |issue=7555 |pages=207–211 |doi=10.1038/nature14317 |pmid=25731166 |pmc=5048219 |arxiv=1502.02783 |bibcode=2015Natur.522..207H |display-authors=1}}</ref><ref>{{cite journal |last1=Lazaridis |first1=I |last2=Patterson |first2=N |date=2014 |title=Ancient human genomes suggest three ancestral populations for present-day Europeans |journal=Nature |volume=513 |issue=7518 |pages=409–413 |doi=10.1038/nature13673 |pmid=25230663 |pmc=4170574 |arxiv=1312.6639 |bibcode=2014Natur.513..409L |display-authors=1}}</ref> Even within historical times, different population groups throughout the world have experienced admixture events that led to their current genetic compositions. For instance, different populations of the Mediterranean, Asia Minor, and around the Arabian Sea exhibit recent historical geneflow from various Sub-Saharan populations, with the most recent influx of Sub-Saharan admixture ranging in date from AD980 to 1754.<ref>{{cite journal |last1=Hellenthal |first1=G |date=2014 |title=A genetic atlas of human admixture history |journal=Science |volume=343 |issue=6172 |pages=750–751|doi=10.1126/science.1243518 |pmid=24531965 |pmc=4209567 |bibcode=2014Sci...343..747H }}</ref> Populations of modern-day Central and Eastern Europe, from Poland in the west and Bulgaria in the south exhibit traces of north-eastern and eastern Asian genetic flow ranging in date from AD440 to 1080 and likely reflect a 'small genetic legacy' of the various Inner Asian nomads, including Huns, Magyars, and Bulgars.<ref>{{cite journal |last1=Hellenthal |first1=G |date=2014 |title=A genetic atlas of human admixture history |journal=Science |volume=343 |issue=6172 |pages=751|doi=10.1126/science.1243518 |pmid=24531965 |pmc=4209567 |bibcode=2014Sci...343..747H }}</ref> |
|||
We nonetheless see wide individual variation in phenotype, which arises from both genetic differences and complex gene-environment interactions. The vast majority of this genetic variation occurs ''within'' groups; very little genetic variation differentiates ''between'' groups.<ref name='Rosenberg2002'/> Crucially, the between-group genetic differences that do exist do not map onto socially recognized categories of race. Furthermore, although human populations show some genetic clustering across geographic space, human genetic variation is "[[Cline (biology)|clinal]]", or continuous.<ref name='AABAstatement'/><ref name='Tishkoff&Kidd2004'/> This, in addition to the fact that different traits vary on different clines, makes it impossible to draw discrete genetic boundaries around human groups. Finally, insights from ancient DNA are revealing that no human population is "pure" – all populations represent a long history of migration and mixing.<ref name=reich>{{cite book |last1=Reich |first1=David |title=Who We Are and How We Got Here: Ancient DNA and the new science of the human past |date=29 March 2018 |publisher=Oxford University Press |isbn=978-0-19-255438-3 |page=xxiv |url=https://books.google.com/books?id=rKZTDwAAQBAJ&pg=PR24 |language=en}}</ref> |
|||
==Sources of human genetic variation== |
==Sources of human genetic variation== |
||
{{Main|Human genetic variation}}Genetic variation arises from [[mutations]], from natural selection, migration between populations ([[gene flow]]) and from the reshuffling of genes through [[sexual reproduction]].<ref>{{Cite journal|last=Livingstone|first=Frank|date=Summer 1962|title=On the Non-Existence of Human Races|url=https://moodle.lse.ac.uk/pluginfile.php/618047/mod_resource/content/0/Livingstone%201962.pdf|journal=Chicago Journals}}</ref> Mutations lead to a change in the DNA structure, as the order of the bases are rearranged. Resultantly, different polypeptide proteins are coded. Some mutations may be positive and can help the individual survive more effectively in their environment. |
{{Main|Human genetic variation}}Genetic variation arises from [[mutations]], from natural selection, migration between populations ([[gene flow]]) and from the reshuffling of genes through [[sexual reproduction]].<ref>{{Cite journal|last=Livingstone|first=Frank|date=Summer 1962|title=On the Non-Existence of Human Races|url=https://moodle.lse.ac.uk/pluginfile.php/618047/mod_resource/content/0/Livingstone%201962.pdf|journal=Chicago Journals|access-date=2019-04-29|archive-date=2021-05-25|archive-url=https://web.archive.org/web/20210525023934/https://moodle.lse.ac.uk/pluginfile.php/618047/mod_resource/content/0/Livingstone%201962.pdf|url-status=dead}}</ref> Mutations lead to a change in the DNA structure, as the order of the bases are rearranged. Resultantly, different polypeptide proteins are coded. Some mutations may be positive and can help the individual survive more effectively in their environment. Mutation is counteracted by [[natural selection]] and by [[genetic drift]]; note too the [[founder effect]], when a small number of initial founders establish a population which hence starts with a correspondingly small degree of genetic variation.<ref>{{Citation|last=Honnay|first=O.|title=Genetic Drift|date=2013 |encyclopedia=Brenner's Encyclopedia of Genetics|pages=251–253|publisher=Elsevier|language=en|doi=10.1016/b978-0-12-374984-0.00616-1 |isbn=978-0-08-096156-9}}</ref> [[Epigenetic inheritance]] involves [[Heredity|heritable]] changes in [[phenotype]] (appearance) or [[gene expression]] caused by mechanisms other than changes in the DNA sequence.<ref>{{Cite journal|last1=Martin|first1=Cyrus|last2=Zhang|first2=Yi|date=June 2007|title=Mechanisms of epigenetic inheritance |journal=Current Opinion in Cell Biology|language=en|volume=19|issue=3|pages=266–272|doi=10.1016/j.ceb.2007.04.002|pmid=17466502}}</ref> |
||
Human phenotypes are highly [[polygenic]] (dependent on interaction by many genes) and are influenced by environment as well as by genetics. |
Human phenotypes are highly [[polygenic]] (dependent on interaction by many genes) and are influenced by environment as well as by genetics. |
||
[[Nucleotide diversity]] is based on single mutations, [[single nucleotide polymorphisms]] (SNPs). The nucleotide diversity between humans is about 0.1 percent (one difference per one thousand [[nucleotide]]s between two humans chosen at random). This amounts to approximately three million SNPs (since the human genome has about three billion nucleotides). There are an estimated ten million SNPs in the human population.<ref>{{ |
[[Nucleotide diversity]] is based on single mutations, [[single nucleotide polymorphisms]] (SNPs). The nucleotide diversity between humans is about 0.1 percent (one difference per one thousand [[nucleotide]]s between two humans chosen at random). This amounts to approximately three million SNPs (since the human genome has about three billion nucleotides). There are an estimated ten million SNPs in the human population.<ref name="Jorde2004">{{cite journal |last1=Jorde |first1=Lynn B |last2=Wooding |first2=Stephen P |author1-link=Lynn Jorde |title=Genetic variation, classification and 'race' |journal=[[Nature Genetics]] |date=November 2004 |volume=36 |issue=11 |series=Supplemental |pages=28–33 |doi=10.1038/ng1435 |pmid=15508000 |url=https://www.nature.com/articles/ng1435 |access-date=27 June 2024 |publisher=[[Nature Portfolio]] |issn=1546-1718 |oclc=8091998144 |s2cid=1500915 |id=Jorde2004 |archive-url=http://web.archive.org/web/20240622010650/https://www.nature.com/articles/ng1435 |archive-date=22 June 2024}}</ref> |
||
Research has shown that non-SNP ([[structural variation|structural]]) variation accounts for more human genetic variation than single nucleotide diversity. Structural variation includes [[copy-number variation]] and results from [[Deletion (genetics)|deletions]], [[Chromosomal inversion|inversions]], [[Genetic insertion|insertions]] and [[Gene duplication|duplications]]. It is estimated that approximately 0.4 to 0.6 percent of the genomes of unrelated people differ.<ref name= |
Research has shown that non-SNP ([[structural variation|structural]]) variation accounts for more human genetic variation than single nucleotide diversity. Structural variation includes [[copy-number variation]] and results from [[Deletion (genetics)|deletions]], [[Chromosomal inversion|inversions]], [[Genetic insertion|insertions]] and [[Gene duplication|duplications]]. It is estimated that approximately 0.4 to 0.6 percent of the genomes of unrelated people differ.<ref name='Tishkoff&Kidd2004'/><ref name="kGP15">{{cite journal|display-authors=6|vauthors=Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR|date=October 2015|title=A global reference for human genetic variation|journal=Nature|volume=526|issue=7571|pages=68–74|bibcode=2015Natur.526...68T|doi=10.1038/nature15393|pmc=4750478|pmid=26432245}}</ref> |
||
== Genetic basis for race == |
== Genetic basis for race == |
||
Much scientific research has been organized around the question of whether or not there is genetic basis for race. In [[Luigi Luca Cavalli-Sforza|Luigi Luca Cavalli-Sforza's]] book (circa 1994) "The History and Geography of Human Genes"<ref name="Cavalli-Sforza1994">{{Cite book |last1=Cavalli-Sforza |first1=Luigi Luca |author-link=Luigi Luca Cavalli-Sforza |url=https://archive.org/details/historygeography0000cava_g9l7 |title=The History and Geography of Human Genes |last2=Menozzi |first2=Paolo |last3=Piazza |first3=Alberto |date=1994 |publisher=Princeton University Press |isbn=978-0-691-08750-4 |location=Princeton |url-access=registration}} |
|||
Much scientific research has been organized around the question of whether or not there is genetic basis for race. According to [[Luigi Luca Cavalli-Sforza]], "From a scientific point of view, the concept of race has failed to obtain any consensus; none is likely, given the gradual variation in existence. It may be objected that the racial stereotypes have a consistency that allows even the layman to classify individuals. However, the major stereotypes, all based on skin color, hair color and form, and facial traits, reflect superficial differences that are not confirmed by deeper analysis with more reliable genetic traits and whose origin dates from recent evolution mostly under the effect of climate and perhaps sexual selection".<ref name="Cavalli-Sforza1994">{{cite book|last1=Cavalli-Sforza|first1=Luigi Luca|url=https://archive.org/details/historygeography0000cava_g9l7|title=The History and Geography of Human Genes|last2=Menozzi|first2=Paolo|last3=Piazza|first3=Alberto|date=1994|publisher=Princeton University Press|isbn=978-0-691-08750-4|location=Princeton|author-link1=Luigi Luca Cavalli-Sforza|url-access=registration|lay-url=https://www.nytimes.com/books/00/08/20/reviews/000820.20ridleyt.html|lay-date=1 December 2013}}</ref><ref>{{cite journal|last=McDonald|first=J|author2=Lehman, DC|date=Spring 2012|title=Forensic DNA analysis|journal=Clinical Laboratory Science|volume=25|issue=2|pages=109–113|doi=10.29074/ascls.25.2.109|pmid=22693781|doi-access=free}}</ref><ref>{{cite web|last=Butler|first=John|title=Genetics and Genomics of Core STR Loci Used in Human Identity Testing*|url=http://www.cstl.nist.gov/div831/strbase/pub_pres/Butler_coreSTRloci_JFS_Mar2006.pdf}}</ref><ref name="Redd 580–5">{{cite journal|last=Redd|first=A. J.|author2=Chamberlain, VF|author3=Kearney, V. F.|author4=Stover, D|author5=Karafet, T.|author6=Calderon, K.|author7=Walsh, B.|author8=Hammer, M. F.|date=May 2006|title=Genetic structure among 38 populations from the United States based on 11 U.S. core Y chromosome STRs.|journal=Journal of Forensic Sciences|volume=51|issue=3|pages=580–585|doi=10.1111/j.1556-4029.2006.00113.x|pmid=16696705|s2cid=23597453}}</ref><ref>{{cite journal|last=Hammer|first=M. F.|author2=Chamberlain, V. F.|author3=Kearney, V. F.|author4=Stover, D.|author5=Zhang, G|author6=Karafet, T.|author7=Walsh, B.|author8=Redd, A. J.|date=December 1, 2006|title=Population structure of Y chromosome SNP haplogroups in the United States and forensic implications for constructing Y chromosome STR databases.|url=http://www.fsijournal.org/article/S0379-0738(05)00615-8/abstract|journal=Forensic Science International|volume=164|issue=1|pages=45–55|doi=10.1016/j.forsciint.2005.11.013|pmid=16337103}}</ref><ref>{{cite journal|last=Sims|first=L. M.|author2=Ballantyne, J.|date=March 2008|title=The golden gene (SLC24A5) differentiates US sub-populations within the ethnically admixed Y-SNP haplogroups.|url=https://www.ncjrs.gov/pdffiles1/nij/grants/211979.pdf|journal=Legal Medicine (Tokyo, Japan)|volume=10|issue=2|pages=72–7|doi=10.1016/j.legalmed.2007.06.004|pmid=17720606}}</ref> |
|||
*{{cite news |author=Mark Ridley |date=August 20, 2000 |title=How Far From the Tree? |type=Review |newspaper=The New York Times |url=https://www.nytimes.com/books/00/08/20/reviews/000820.20ridleyt.html |access-date=March 3, 2017 |archive-date=March 17, 2017 |archive-url=https://web.archive.org/web/20170317224500/http://www.nytimes.com/books/00/08/20/reviews/000820.20ridleyt.html |url-status=live }}</ref> he writes, "From a scientific point of view, the concept of race has failed to obtain any consensus; none is likely, given the gradual variation in existence. It may be objected that the racial stereotypes have a consistency that allows even the layman to classify individuals. However, the major stereotypes, all based on skin color, hair color and form, and facial traits, reflect superficial differences that are not confirmed by deeper analysis with more reliable genetic traits and whose origin dates from recent evolution mostly under the effect of climate and perhaps sexual selection". |
|||
In 2018 geneticist [[David Reich (geneticist)|David Reich]] reaffirmed the conclusion that the traditional views which assert a biological basis for race are wrong: |
|||
{{Blockquote|text=Today, many people assume that humans can be grouped biologically into "primeval" groups, corresponding to our notion of "races"... But this long-held view about "race" has just in the last years been proven wrong.|author=David Reich|title=Who We Are and How We Got Here|source=(Introduction, pg. xxiv).}} |
|||
In 1956, some scientists proposed that race may be similar to dog breeds within dogs. However, this theory has since been discarded, with one of the main reasons being that purebred dogs have been specifically bred artificially, whereas human races developed organically.<ref>{{Cite journal |last1=Norton |first1=Heather L. |last2=Quillen |first2=Ellen E. |last3=Bigham |first3=Abigail W. |last4=Pearson |first4=Laurel N. |last5=Dunsworth |first5=Holly |date=2019-07-09 |title=Human races are not like dog breeds: refuting a racist analogy |journal=Evolution: Education and Outreach |volume=12 |issue=1 |pages=17 |doi=10.1186/s12052-019-0109-y |issn=1936-6434 |doi-access=free}}</ref> Furthermore, the genetic variation between purebred dog breeds is far greater than that of human populations. Dog-breed intervariation is roughly 27.5%, whereas human populations inter-variation is only at 10-15.6%.<ref>{{cite journal |last1=Templeton |first1=Alan |title=Human Races: A Genetic and Evolutionary Perspective |journal=American Anthropologist |date=2008 |volume=100 |issue=3 |page=633 |doi=10.1525/aa.1998.100.3.632 |url=https://anthrosource-onlinelibrary-wiley-com.proxy2.cl.msu.edu/doi/full/10.1525/aa.1998.100.3.632 |access-date=19 July 2024}}</ref><ref>{{Cite web |first=Elaine A. |last=Ostrander |date=2017-02-06 |title=Genetics and the Shape of Dogs |url=https://www.americanscientist.org/article/genetics-and-the-shape-of-dogs |access-date=2024-03-12 |website=American Scientist |language=en}}</ref><ref>{{Cite journal |last=Bamshad |first=Michael |date=2005 |title=Genetic Influenes on Health: Does Race Matter? |url=https://pubmed.ncbi.nlm.nih.gov/16118384/ |journal=JAMA |volume=294 |issue=8 |page=938 |doi=10.1001/jama.294.8.937 |pmid=16118384 |access-date=18 July 2024}}</ref><ref>{{Cite journal |last1=Parker |first1=Heidi G. |last2=Kim |first2=Lisa V. |last3=Sutter |first3=Nathan B. |last4=Carlson |first4=Scott |last5=Lorentzen |first5=Travis D. |last6=Malek |first6=Tiffany B. |last7=Johnson |first7=Gary S. |last8=DeFrance |first8=Hawkins B. |last9=Ostrander |first9=Elaine A. |last10=Kruglyak |first10=Leonid |date=2004-05-21 |title=Genetic Structure of the Purebred Domestic Dog |url=https://www.science.org/doi/10.1126/science.1097406 |journal=Science |language=en |volume=304 |issue=5674 |pages=1160–1164 |doi=10.1126/science.1097406 |issn=0036-8075 |pmid=15155949 |bibcode=2004Sci...304.1160P |access-date=18 July 2024}}</ref> Including non purebreds would substantially decrease the 27.5% genetic variance, however. Mammal taxonomy is rarely defined by genetic variance alone. |
|||
=={{anchor|Methods in human ancestry and population genetic structure research}}Research methods== |
=={{anchor|Methods in human ancestry and population genetic structure research}}Research methods== |
||
| Line 34: | Line 48: | ||
==={{anchor|Visible traits, proteins, and genes studied}}Early studies of traits, proteins, and genes=== |
==={{anchor|Visible traits, proteins, and genes studied}}Early studies of traits, proteins, and genes=== |
||
{{See also|Race (classification of human beings)}} |
{{See also|Race (classification of human beings)}} |
||
Early racial classification attempts measured [[anthropometry|surface traits]], particularly skin color, hair color and texture, eye color, and head size and shape. (Measurements of the latter through [[craniometry]] were repeatedly discredited in the late 19th and mid-20th centuries due to a lack of correlation of phenotypic traits with racial categorization.<ref name="Orsucci">Andrea Orsucci |
Early racial classification attempts measured [[anthropometry|surface traits]], particularly skin color, hair color and texture, eye color, and head size and shape. (Measurements of the latter through [[craniometry]] were repeatedly discredited in the late 19th and mid-20th centuries due to a lack of correlation of phenotypic traits with racial categorization.<ref name="Orsucci">{{cite web |first=Andrea |last=Orsucci |url=http://www.unifi.it/riviste/cromohs/3_98/orsucci.html |title=Ariani, indogermani, stirpi mediterranee: aspetti del dibattito sulle razze europee (1870–1914) |archive-url=https://archive.today/20121218231754/http://www.unifi.it/riviste/cromohs/3_98/orsucci.html|archive-date=December 18, 2012 |work=Cromohs |year=1998 |lang=it}}</ref>) In actuality, biological adaptation plays the biggest role in these bodily features and skin type. A relative handful of genes accounts for the inherited factors shaping a person's appearance.<ref>{{cite news |first=Natalie |last=Angier |url=http://partners.nytimes.com/library/national/science/082200sci-genetics-race.html|title=Do Races Differ? Not Really, DNA Shows|date=22 August 2000|work=The New York Times|access-date=3 September 2011|archive-date=30 April 2021|archive-url=https://web.archive.org/web/20210430051310/https://archive.nytimes.com/www.nytimes.com/library/national/science/082200sci-genetics-race.html|url-status=live}}</ref><ref>{{Cite journal|last1=Owens|first1=Kelly|last2=King|first2=Mary-Claire|date=1999-10-15|title=Genomic Views of Human History|journal=Science|volume=286|issue=5439|pages=451–453|doi=10.1126/science.286.5439.451|issn=0036-8075|pmid=10521333|quote=Variation in other traits popularly used to identify 'races' is likely to be due to similarly straightforward mechanisms, involving limited numbers of genes with very specific physiological effects.}}</ref> Humans have an estimated 19,000–20,000 human protein-coding genes.<ref>{{cite journal|date=November 2014|title=Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes|journal=Human Molecular Genetics|volume=23|issue=22|pages=5866–5878|doi=10.1093/hmg/ddu309|pmc=4204768|pmid=24939910|vauthors=Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML}}</ref> Richard Sturm and David Duffy describe 11 genes that affect skin pigmentation and explain most variations in [[human skin color]], the most significant of which are [[Melanocortin 1 receptor|MC1R]], ASIP, [[OCA2]], and TYR.<ref>{{Cite journal|last1=Sturm|first1=Richard A.|last2=Duffy|first2=David L.|date=2012|title=Human pigmentation genes under environmental selection|journal=Genome Biology|volume=13|issue=9|pages=248|doi=10.1186/gb-2012-13-9-248|issn=1474-760X|pmc=3491390|pmid=23110848 |doi-access=free }}</ref> There is evidence that as many as 16 different genes could be responsible for [[eye color]] in humans; however, the main two genes associated with eye color variation are ''[[OCA2]]'' and ''[[HERC2]]'', and both are localized in chromosome 15.<ref name="nature.com">{{cite journal|last2=Rabago-Smith|first2=Montserrat|title=Genotype-phenotype associations and human eye color|journal=[[Journal of Human Genetics]]|date=January 2011|volume=56|issue=1|pages=5–7|doi=10.1038/jhg.2010.126|pmid=20944644|last1=White|first1=Désirée|doi-access=free}}</ref> |
||
==== Analysis of blood proteins and between-group genetics ==== |
==== Analysis of blood proteins and between-group genetics ==== |
||
[[File:groupa.png|thumb|right|upright=1.4|alt=Multicolored world map|Geographic distribution of blood group A]] |
[[File:groupa.png|thumb|right|upright=1.4|alt=Multicolored world map|Geographic distribution of blood group A]] |
||
[[File:groupb.png|thumb|right|upright=1.4|alt=Multicolored world map|Geographic distribution of blood group B]] |
[[File:groupb.png|thumb|right|upright=1.4|alt=Multicolored world map|Geographic distribution of blood group B]] |
||
Before the discovery of DNA, scientists used blood proteins (the [[human blood group systems]]) to study human genetic variation. Research by [[Ludwik and Hanka Herschfeld]] during [[World War I]] found that the incidence of [[blood groups]] A and B differed by region; for example, among Europeans 15 percent were group B and 40 percent group A. Eastern Europeans and Russians had a higher incidence of group B; people from India had the greatest incidence. The Herschfelds concluded that humans comprised two "biochemical races", originating separately. It was hypothesized that these two races later mixed, resulting in the patterns of groups A and B. This was one of the first theories of racial differences to include the idea that human variation did not correlate with genetic variation. It was expected that groups with similar proportions of blood groups would be more closely related, but instead it was often found that groups separated by great distances (such as those from Madagascar and Russia), had similar incidences.<ref name="bryansykes">{{cite book | |
Before the discovery of DNA, scientists used blood proteins (the [[human blood group systems]]) to study human genetic variation. Research by [[Ludwik and Hanka Herschfeld]] during [[World War I]] found that the incidence of [[blood groups]] A and B differed by region; for example, among Europeans 15 percent were group B and 40 percent group A. Eastern Europeans and Russians had a higher incidence of group B; people from India had the greatest incidence. The Herschfelds concluded that humans comprised two "biochemical races", originating separately. It was hypothesized that these two races later mixed, resulting in the patterns of groups A and B. This was one of the first theories of racial differences to include the idea that human variation did not correlate with genetic variation. It was expected that groups with similar proportions of blood groups would be more closely related, but instead it was often found that groups separated by great distances (such as those from Madagascar and Russia), had similar incidences.<ref name="bryansykes">{{cite book |last=Sykes |first=Bryan |chapter=From Blood Groups to Genes |title=The seven daughters of Eve |publisher=Norton |location=New York |year=2001 |pages=[https://archive.org/details/sevendaughtersof00syke/page/32 32]–51 |isbn=978-0-393-02018-2 |chapter-url-access=registration |chapter-url=https://archive.org/details/sevendaughtersof00syke }}</ref> It was later discovered that the [[ABO blood group system]] is not just common to humans, but shared with other primates,<ref>{{Cite book|title=Molecular biology and evolution of blood group and MHC antigens in primates|last2=Klein|first2=Jan|last3=Socha|first3=Wladyslaw W.|date=2012|publisher=Springer Science & Business Media|isbn=978-3-642-59086-3|last1=Blancher|first1=Antoine}}</ref> and likely predates all human groups.<ref>{{Cite journal|last2=Thompson|first2=Emma E.|last3=Flutre|first3=Timothée|last4=Lovstad|first4=Jessica|last5=Venkat|first5=Aarti|last6=Margulis|first6=Susan W.|last7=Moyse|first7=Jill|last8=Ross|first8=Steve|last9=Gamble|first9=Kathryn|date=2012-11-06|title=The ABO blood group is a trans-species polymorphism in primates|journal=Proceedings of the National Academy of Sciences|volume=109|issue=45|pages=18493–18498|doi=10.1073/pnas.1210603109|issn=0027-8424|pmid=23091028|last1=Ségurel|first1=Laure|last10=Sella|first10=Guy|last11=Ober|first11=Carole|last12=Przeworski|first12=Molly|pmc=3494955|arxiv=1208.4613|bibcode=2012PNAS..10918493S|doi-access=free}}</ref> |
||
In 1972, [[Richard Lewontin]] performed a F<sub>ST</sub> statistical analysis using 17 markers (including blood-group proteins). He found that the majority of genetic differences between humans (85.4 percent) were found within a population, 8.3 percent were found between populations within a race and 6.3 percent were found to differentiate races (Caucasian, African, Mongoloid, South Asian Aborigines, Amerinds, Oceanians, and Australian Aborigines in his study). Since then, other analyses have found F<sub>ST</sub> values of 6–10 percent between continental human groups, 5–15 percent between different populations on the same continent and 75–85 percent within populations.<ref>{{cite book|last=Lewontin|first=Richard|title=Evolutionary Biology|date=1972|isbn=978-1-4684-9065-7|editor1=Theodosius Dobzhansky|volume=6|pages=381–398|chapter=The Apportionment of Human Diversity|doi=10.1007/978-1-4684-9063-3_14|author-link=Richard Lewontin|editor2=Max K. Hecht|editor3=William C. Steere}}</ref><ref name="Risch2002">{{cite journal|last1=Risch|first1=Neil|last2=Burchard|first2=Esteban|last3=Ziv|first3=Elad|last4=Tang|first4=Hua|year=2002|title=Categorization of humans in biomedical research: genes, race and disease|journal=Genome Biology|volume=3|issue=7|pages=comment2007.1|doi=10.1186/gb-2002-3-7-comment2007|issn=1465-6906|pmc=139378|pmid=12184798}}</ref><ref name="Templeton1998">{{cite book|last=Templeton|first=Alan R.|title=Genetic nature/culture: anthropology and science beyond the two-culture divide|publisher=University of California Press|year=2003|isbn=978-0-520-23792-6|editor1-last=Goodman|editor1-first=Alan H.|location=Berkeley|pages=234–257|chapter=Human Races in the Context of Recent Human Evolution: A Molecular Genetic Perspective|access-date=23 September 2014|editor2-last=Heath|editor2-first=Deborah|editor3-last=Lindee|editor3-first=M. Susan|chapter-url=http://www.ucpress.edu/book.php?isbn=9780520237933}}</ref><ref>{{cite journal|vauthors=Ossorio P, Duster T|date=January 2005|title=Race and genetics: controversies in biomedical, behavioral, and forensic sciences|journal=The American Psychologist|volume=60|issue=1|pages=115–128|doi=10.1037/0003-066X.60.1.115|pmid=15641926}}</ref><ref name="Lewontin2005">Lewontin |
In 1972, [[Richard Lewontin]] performed a F<sub>ST</sub> statistical analysis using 17 markers (including blood-group proteins). He found that the majority of genetic differences between humans (85.4 percent) were found within a population, 8.3 percent were found between populations within a race and 6.3 percent were found to differentiate races (Caucasian, African, Mongoloid, South Asian Aborigines, Amerinds, Oceanians, and Australian Aborigines in his study). Since then, other analyses have found F<sub>ST</sub> values of 6–10 percent between continental human groups, 5–15 percent between different populations on the same continent and 75–85 percent within populations.<ref>{{cite book|last=Lewontin|first=Richard|title=Evolutionary Biology|date=1972|isbn=978-1-4684-9065-7|editor1=Theodosius Dobzhansky|volume=6|pages=381–398|chapter=The Apportionment of Human Diversity|doi=10.1007/978-1-4684-9063-3_14|s2cid=21095796 |author-link=Richard Lewontin|editor2=Max K. Hecht|editor3=William C. Steere}}</ref><ref name="Risch2002">{{cite journal|last1=Risch|first1=Neil|last2=Burchard|first2=Esteban|last3=Ziv|first3=Elad|last4=Tang|first4=Hua|year=2002|title=Categorization of humans in biomedical research: genes, race and disease|journal=Genome Biology|volume=3|issue=7|pages=comment2007.1|doi=10.1186/gb-2002-3-7-comment2007|issn=1465-6906|pmc=139378|pmid=12184798 |doi-access=free }}</ref><ref name="Templeton1998">{{cite book|last=Templeton|first=Alan R.|title=Genetic nature/culture: anthropology and science beyond the two-culture divide|publisher=University of California Press|year=2003|isbn=978-0-520-23792-6|editor1-last=Goodman|editor1-first=Alan H.|location=Berkeley|pages=234–257|chapter=Human Races in the Context of Recent Human Evolution: A Molecular Genetic Perspective|access-date=23 September 2014|editor2-last=Heath|editor2-first=Deborah|editor3-last=Lindee|editor3-first=M. Susan|chapter-url=http://www.ucpress.edu/book.php?isbn=9780520237933|archive-date=9 November 2014|archive-url=https://web.archive.org/web/20141109142919/http://www.ucpress.edu/book.php?isbn=9780520237933|url-status=live}}</ref><ref>{{cite journal|vauthors=Ossorio P, Duster T|date=January 2005|title=Race and genetics: controversies in biomedical, behavioral, and forensic sciences|journal=The American Psychologist|volume=60|issue=1|pages=115–128|doi=10.1037/0003-066X.60.1.115|pmid=15641926}}</ref><ref name="Lewontin2005">{{cite web |last=Lewontin |first=R. C. |year=2005 |url=http://raceandgenomics.ssrc.org/Lewontin/ |title=Confusions About Human Races |archive-url=https://web.archive.org/web/20130504132215/http://raceandgenomics.ssrc.org/Lewontin/ |archive-date=2013-05-04 |department=Race and Genomics |website=Social Sciences Research Council |access-date=28 December 2006}}</ref> This view has been affirmed by the [[American Anthropological Association]] and the American Association of Physical Anthropologists since.<ref name=":0">{{Cite journal|last1=Long|first1=Jeffrey C.|last2=Kittles|first2=Rick A.|date=2009|title=Human Genetic Diversity and the Nonexistence of Biological Races|url=https://muse.jhu.edu/journals/human_biology/v081/81.5-6.long.html|journal=Human Biology|volume=81|issue=5|pages=777–798|doi=10.3378/027.081.0621|issn=1534-6617|pmid=20504196|access-date=2016-01-13|s2cid=30709062|archive-date=2020-03-13|archive-url=https://web.archive.org/web/20200313181641/https://muse.jhu.edu/article/381883|url-status=live}}</ref> |
||
==== Critiques of blood protein analysis ==== |
==== Critiques of blood protein analysis ==== |
||
While acknowledging Lewontin's observation that humans are genetically homogeneous, [[A. W. F. Edwards]] in his 2003 paper "[[Lewontin's Fallacy|Human Genetic Diversity: Lewontin's Fallacy]]" argued that information distinguishing populations from each other is hidden in the correlation structure of allele frequencies, making it possible to classify individuals using mathematical techniques. Edwards argued that even if the probability of misclassifying an individual based on a single genetic marker is as high as 30 percent (as Lewontin reported in 1972), the misclassification probability nears zero if enough genetic markers are studied simultaneously. Edwards saw Lewontin's argument as based on a political stance, denying biological differences to argue for social equality.<ref name="Edwards2003">{{cite journal|author=Edwards AW|date=August 2003|title=Human genetic diversity: Lewontin's fallacy|journal=BioEssays|volume=25|issue=8|pages=798–801|doi=10.1002/bies.10315|pmid=12879450}}</ref> Edwards' paper is reprinted, commented upon by experts such as [[Noah Rosenberg]], and given further context in an interview with philosopher of science Rasmus Grønfeldt Winther in a recent anthology.<ref>{{cite book|author-last=Winther|author-first=Rasmus Grønfeldt|url=https://www.cambridge.org/de/academic/subjects/life-sciences/genetics/phylogenetic-inference-selection-theory-and-history-science-selected-papers-w-f-edwards-commentaries|title=Phylogenetic Inference, Selection Theory, and History of Science: Selected Papers of A. W. F. Edwards with Commentaries|date=2018|publisher=[[Cambridge University Press]]|isbn=9781107111721|location=Cambridge, U.K.}}</ref> |
While acknowledging Lewontin's observation that humans are genetically homogeneous, [[A. W. F. Edwards]] in his 2003 paper "[[Lewontin's Fallacy|Human Genetic Diversity: Lewontin's Fallacy]]" argued that information distinguishing populations from each other is hidden in the correlation structure of allele frequencies, making it possible to classify individuals using mathematical techniques. Edwards argued that even if the probability of misclassifying an individual based on a single genetic marker is as high as 30 percent (as Lewontin reported in 1972), the misclassification probability nears zero if enough genetic markers are studied simultaneously. Edwards saw Lewontin's argument as based on a political stance, denying biological differences to argue for social equality.<ref name="Edwards2003">{{cite journal|author=Edwards AW|date=August 2003|title=Human genetic diversity: Lewontin's fallacy|journal=BioEssays|volume=25|issue=8|pages=798–801|doi=10.1002/bies.10315|pmid=12879450}}</ref> Edwards' paper is reprinted, commented upon by experts such as [[Noah Rosenberg]], and given further context in an interview with philosopher of science Rasmus Grønfeldt Winther in a recent anthology.<ref>{{cite book|author-last=Winther|author-first=Rasmus Grønfeldt|url=https://www.cambridge.org/de/academic/subjects/life-sciences/genetics/phylogenetic-inference-selection-theory-and-history-science-selected-papers-w-f-edwards-commentaries|title=Phylogenetic Inference, Selection Theory, and History of Science: Selected Papers of A. W. F. Edwards with Commentaries|date=2018|publisher=[[Cambridge University Press]]|isbn=9781107111721|location=Cambridge, U.K.|access-date=2018-12-13|archive-date=2019-08-15|archive-url=https://web.archive.org/web/20190815031238/https://www.cambridge.org/de/academic/subjects/life-sciences/genetics/phylogenetic-inference-selection-theory-and-history-science-selected-papers-w-f-edwards-commentaries|url-status=live}}</ref> |
||
As referred to before, Edwards criticises Lewontin's paper as he took 17 different traits and analysed them independently, without looking at them in conjunction with any other protein. Thus, it would have been fairly convenient for Lewontin to come up with the conclusion that racial naturalism is not tenable, according to his argument.<ref>{{cite book|last1=Edwards|first1=AWF|title=Human genetic diversity: Lewontin's fallacy, BioEssays|date=2003|pages=798–801}}</ref> Sesardic also strengthened Edwards' view, as he used an illustration referring to squares and triangles, and showed that if you look at one trait in isolation, then it will most likely be a bad predicator of which group the individual belongs to.<ref>{{cite book|last1=Sesardic|first1=N.|title=Race: a social destruction of a biological concept. Biology and Philosophy|date=2010|pages=143–162}}</ref> In contrast, in a 2014 paper, reprinted in the 2018 Edwards Cambridge University Press volume, Rasmus Grønfeldt Winther argues that "Lewontin's Fallacy" is effectively a misnomer, as there really are two different sets of methods and questions at play in studying the genomic population structure of our species: "variance partitioning" and "clustering analysis." According to Winther, they are "two sides of the same mathematics coin" and neither "necessarily implies anything about the ''reality'' of human groups."<ref>{{Cite book | last1 = Winther | first1 = R.G. | chapter = The Genetic Reification of "Race"? A Story of Two Mathematical Methods | title = Phylogenetic Inference, Selection Theory, and History of Science: Selected Papers of AWF Edwards with Commentaries | editor = R.G. Winther | pages = 489, 488–508 | year = 2018 | isbn = 9781107111721 |url=https://www.cambridge.org/de/academic/subjects/life-sciences/genetics/phylogenetic-inference-selection-theory-and-history-science-selected-papers-w-f-edwards-commentaries}}</ref> |
As referred to before, Edwards criticises Lewontin's paper as he took 17 different traits and analysed them independently, without looking at them in conjunction with any other protein. Thus, it would have been fairly convenient for Lewontin to come up with the conclusion that racial naturalism is not tenable, according to his argument.<ref>{{cite book|last1=Edwards|first1=AWF|title=Human genetic diversity: Lewontin's fallacy, BioEssays|date=2003|pages=798–801}}</ref> Sesardic also strengthened Edwards' view, as he used an illustration referring to squares and triangles, and showed that if you look at one trait in isolation, then it will most likely be a bad predicator of which group the individual belongs to.<ref>{{cite book|last1=Sesardic|first1=N.|title=Race: a social destruction of a biological concept. Biology and Philosophy|date=2010|pages=143–162}}</ref> In contrast, in a 2014 paper, reprinted in the 2018 Edwards Cambridge University Press volume, Rasmus Grønfeldt Winther argues that "Lewontin's Fallacy" is effectively a misnomer, as there really are two different sets of methods and questions at play in studying the genomic population structure of our species: "variance partitioning" and "clustering analysis." According to Winther, they are "two sides of the same mathematics coin" and neither "necessarily implies anything about the ''reality'' of human groups."<ref>{{Cite book | last1 = Winther | first1 = R.G. | chapter = The Genetic Reification of "Race"? A Story of Two Mathematical Methods | title = Phylogenetic Inference, Selection Theory, and History of Science: Selected Papers of AWF Edwards with Commentaries | editor = R.G. Winther | pages = 489, 488–508 | year = 2018 | publisher = Cambridge University Press | isbn = 9781107111721 | url = https://www.cambridge.org/de/academic/subjects/life-sciences/genetics/phylogenetic-inference-selection-theory-and-history-science-selected-papers-w-f-edwards-commentaries | access-date = 2018-12-13 | archive-date = 2019-08-15 | archive-url = https://web.archive.org/web/20190815031238/https://www.cambridge.org/de/academic/subjects/life-sciences/genetics/phylogenetic-inference-selection-theory-and-history-science-selected-papers-w-f-edwards-commentaries | url-status = live }}</ref> |
||
==={{anchor|Population genetic structure and genetic distance}}Current studies of population genetics=== |
==={{anchor|Population genetic structure and genetic distance}}Current studies of population genetics=== |
||
{{Update|section|reason=All references are more than 13 years old|date=December 2022}} |
|||
Researchers currently use [[genetic testing]], which may involve hundreds (or thousands) of [[genetic marker]]s or the entire genome. |
Researchers currently use [[genetic testing]], which may involve hundreds (or thousands) of [[genetic marker]]s or the entire genome. |
||
===={{anchor|Population genetic structure}}Structure==== |
===={{anchor|Population genetic structure}}Structure==== |
||
[[File:Principle component analysis of Levantine populations.png|thumb|Principal component analysis of fifty populations, color-coded by region, illustrates the differentiation and overlap of populations found using this method of analysis.]] |
[[File:Principle component analysis of Levantine populations.png|thumb|Principal component analysis of fifty populations, color-coded by region, illustrates the differentiation and overlap of populations found using this method of analysis.]] |
||
[[File:Human genetic variant counts by region.svg|thumb|Individuals mostly have genetic variants which are found in multiple regions of the world. |
[[File:Human genetic variant counts by region.svg|thumb|Individuals mostly have genetic variants which are found in multiple regions of the world. Based on data from "A unified genealogy of modern and ancient genomes".<ref>{{cite bioRxiv |last1=Wohns |first1=Anthony Wilder |last2=Wong | first2=Yan |last3=Jeffery |first3=Ben |last4=Akbari |first4=Ali |last5=Mallick |first5=Swapan |last6=Pinhasi |first6=Ron |last7=Patterson |first7=Nick |last8=Reich |first8=David |last9=Kelleher |first9=Jerome |last10=McVean |first10=Gil |date=April 15, 2021 |title=A unified genealogy of modern and ancient genomes |biorxiv=10.1101/2021.02.16.431497}}</ref>]] |
||
Several methods to examine and quantify genetic subgroups exist, including [[cluster analysis|cluster]] and [[principal components analysis]]. Genetic markers from individuals are examined to find a population's genetic structure. While subgroups overlap when examining variants of one marker only, when a number of markers are examined different subgroups have different average genetic structure. An individual may be described as belonging to several subgroups. These subgroups may be more or less distinct, depending on how much overlap there is with other subgroups.<ref name="Witherspoon">{{cite journal |last1=Witherspoon |first1=D. J. |last2=Wooding |first2=S. |last3=Rogers |first3=A. R. |last4=Marchani |first4=E. E. |last5=Watkins |first5=W. S. |last6=Batzer |first6=M. A. |last7=Jorde |first7=L. B. |title=Genetic Similarities Within and Between Human Populations |journal=Genetics |volume=176 |issue=1 |date=2007 |pages=351–359 |issn=0016-6731 |pmid=17339205 |pmc=1893020 |doi=10.1534/genetics.106.067355 }}</ref> |
Several methods to examine and quantify genetic subgroups exist, including [[cluster analysis|cluster]] and [[principal components analysis]]. Genetic markers from individuals are examined to find a population's genetic structure. While subgroups overlap when examining variants of one marker only, when a number of markers are examined different subgroups have different average genetic structure. An individual may be described as belonging to several subgroups. These subgroups may be more or less distinct, depending on how much overlap there is with other subgroups.<ref name="Witherspoon">{{cite journal |last1=Witherspoon |first1=D. J. |last2=Wooding |first2=S. |last3=Rogers |first3=A. R. |last4=Marchani |first4=E. E. |last5=Watkins |first5=W. S. |last6=Batzer |first6=M. A. |last7=Jorde |first7=L. B. |title=Genetic Similarities Within and Between Human Populations |journal=Genetics |volume=176 |issue=1 |date=2007 |pages=351–359 |issn=0016-6731 |pmid=17339205 |pmc=1893020 |doi=10.1534/genetics.106.067355 }}</ref> |
||
| Line 59: | Line 74: | ||
The results obtained from cluster analyses depend on several factors: |
The results obtained from cluster analyses depend on several factors: |
||
* A large number genetic markers studied facilitates finding distinct clusters.<ref name=Tang2005>{{cite journal |
* A large number of genetic markers studied facilitates finding distinct clusters.<ref name=Tang2005>{{cite journal |vauthors=Tang H, Quertermous T, Rodriguez B, etal |title=Genetic Structure, Self-Identified Race/Ethnicity, and Confounding in Case-Control Association Studies |journal=American Journal of Human Genetics |volume=76 |issue=2 |pages=268–75 |date=February 2005 |pmid=15625622 |pmc=1196372 |doi=10.1086/427888}}</ref> |
||
* Some genetic markers vary more than others, so fewer are required to find distinct clusters.<ref name= |
* Some genetic markers vary more than others, so fewer are required to find distinct clusters.<ref name='Rosenberg2002'/> [[Ancestry-informative marker]]s exhibit substantially different frequencies between populations from different geographical regions. Using AIMs, scientists can determine a person's ancestral continent of origin based solely on their DNA. AIMs can also be used to determine someone's admixture proportions.<ref>{{cite web| last = Lewontin| first = R. C.| author-link = Richard Lewontin| title = Confusions About Human Races| url = http://raceandgenomics.ssrc.org/Lewontin/| access-date = 2007-01-09| archive-date = 2013-05-04| archive-url = https://web.archive.org/web/20130504132215/http://raceandgenomics.ssrc.org/Lewontin/| url-status = live}}</ref> |
||
* The more individuals studied, the easier it becomes to detect distinct clusters ([[Errors and residuals in statistics|statistical noise]] is reduced).<ref name="Rosenberg2002"/> |
* The more individuals studied, the easier it becomes to detect distinct clusters ([[Errors and residuals in statistics|statistical noise]] is reduced).<ref name="Rosenberg2002"/> |
||
* Low genetic variation makes it more difficult to find distinct clusters.<ref name="Rosenberg2002"/> Greater geographic distance generally increases genetic variation, making identifying clusters easier.<ref>{{cite journal |vauthors=Kittles RA, Weiss KM |title=Race, ancestry, and genes: implications for defining disease risk |journal=Annual Review of Genomics and Human Genetics |volume=4 |pages=33–67 |year=2003 |pmid=14527296 |doi=10.1146/annurev.genom.4.070802.110356}}</ref> |
* Low genetic variation makes it more difficult to find distinct clusters.<ref name="Rosenberg2002"/> Greater geographic distance generally increases genetic variation, making identifying clusters easier.<ref>{{cite journal |vauthors=Kittles RA, Weiss KM |title=Race, ancestry, and genes: implications for defining disease risk |journal=Annual Review of Genomics and Human Genetics |volume=4 |pages=33–67 |year=2003 |pmid=14527296 |doi=10.1146/annurev.genom.4.070802.110356}}</ref> |
||
*A similar cluster structure is seen with different genetic markers when the number of genetic markers included is sufficiently large. The clustering structure obtained with different statistical techniques is similar. A similar cluster structure is found in the original sample with a [[Sampling (statistics)|subsample]] of the original sample.<ref name="Rosenberg2005">{{cite journal|author1=Rosenberg NA |author2=Mahajan S |author3=Ramachandran S |author4=Zhao C |author-link5=Jonathan K. Pritchard |author5=Pritchard JK |author6=Feldman MW|date=December 2005|title=Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure|journal=PLOS Genetics|volume=1|issue=6|pages=e70|doi=10.1371/journal.pgen.0010070|pmc=1310579|pmid=16355252}}</ref> |
*A similar cluster structure is seen with different genetic markers when the number of genetic markers included is sufficiently large. The clustering structure obtained with different statistical techniques is similar. A similar cluster structure is found in the original sample with a [[Sampling (statistics)|subsample]] of the original sample.<ref name="Rosenberg2005">{{cite journal|author1=Rosenberg NA |author2=Mahajan S |author3=Ramachandran S |author4=Zhao C |author-link5=Jonathan K. Pritchard |author5=Pritchard JK |author6=Feldman MW|date=December 2005|title=Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure|journal=PLOS Genetics|volume=1|issue=6|pages=e70|doi=10.1371/journal.pgen.0010070|pmc=1310579|pmid=16355252 |doi-access=free }}</ref> |
||
Recent studies have been published using an increasing number of genetic markers.<ref name="Rosenberg2002" /><ref name="Rosenberg2005" /><ref>{{Cite journal|last1=Li|first1=J. Z.|last2=Absher|first2=D. M.|last3=Tang|first3=H.|last4=Southwick|first4=A. M.|last5=Casto|first5=A. M.|last6=Ramachandran|first6=S.|last7=Cann|first7=H. M.|last8=Barsh|first8=G. S.|last9=Feldman|first9=M.|year=2008|title=Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation|journal=Science|volume=319|issue=5866|pages=1100–1104|bibcode=2008Sci...319.1100L|doi=10.1126/science.1153717|pmid=18292342|last10=Cavalli-Sforza|first10=L. L.|last11=Myers|first11=R. M.|s2cid=53541133}}</ref><ref>{{Cite journal|last1=Jakobsson|first1=M.|last2=Scholz|first2=S. W.|last3=Scheet|first3=P.|last4=Gibbs|first4=J. R.|last5=Vanliere|first5=J. M.|last6=Fung|first6=H. C.|last7=Szpiech|first7=Z. A.|last8=Degnan|first8=J. H.|last9=Wang|first9=K.|year=2008|title=Genotype, haplotype and copy-number variation in worldwide human populations|journal=Nature|volume=451|issue=7181|pages=998–1003|bibcode=2008Natur.451..998J|doi=10.1038/nature06742|pmid=18288195|last10=Guerreiro|first10=R.|last11=Bras|first11=J. M.|last12=Schymick|first12=J. C.|last13=Hernandez|first13=D. G.|last14=Traynor|first14=B. J.|last15=Simon-Sanchez|first15=J.|last16=Matarin|first16=M.|last17=Britton|first17=A.|last18=Van De Leemput|first18=J.|last19=Rafferty|first19=I.|last20=Bucan|first20=M.|last21=Cann|first21=H. M.|last22=Hardy|first22=J. A.|last23=Rosenberg|first23=N. A.|last24=Singleton|first24=A. B.|hdl=2027.42/62552|s2cid=11074384|url=https://deepblue.lib.umich.edu/bitstream/2027.42/62552/1/nature06742.pdf|hdl-access=free}}</ref><ref>{{Cite journal|last1=Xing|first1=J.|last2=Watkins|first2=W. S.|last3=Witherspoon|first3=D. J.|last4=Zhang|first4=Y.|last5=Guthery|first5=S. L.|last6=Thara|first6=R.|last7=Mowry|first7=B. J.|last8=Bulayeva|first8=K.|last9=Weiss|first9=R. B.|year=2009|title=Fine-scaled human genetic structure revealed by SNP microarrays|journal=Genome Research|volume=19|issue=5|pages=815–825|doi=10.1101/gr.085589.108|pmc=2675970|pmid=19411602|last10=Jorde|first10=L. B.}}</ref><ref>{{Cite journal|last1=López Herráez|first1=D.|last2=Bauchet|first2=M.|last3=Tang|first3=K.|last4=Theunert|first4=C.|last5=Pugach|first5=I.|last6=Li|first6=J.|last7=Nandineni|first7=M. R.|last8=Gross|first8=A.|last9=Scholz|first9=M.|year=2009|editor1-last=Hawks|editor1-first=John|title=Genetic Variation and Recent Positive Selection in Worldwide Human Populations: Evidence from Nearly 1 Million SNPs|journal=PLOS ONE|volume=4|issue=11|pages=e7888|bibcode=2009PLoSO...4.7888L|doi=10.1371/journal.pone.0007888|pmc=2775638|pmid=19924308|last10=Stoneking|first10=M.|doi-access=free}}</ref> |
Recent studies have been published using an increasing number of genetic markers.<ref name="Rosenberg2002" /><ref name="Rosenberg2005" /><ref>{{Cite journal|last1=Li|first1=J. Z.|last2=Absher|first2=D. M.|last3=Tang|first3=H.|last4=Southwick|first4=A. M.|last5=Casto|first5=A. M.|last6=Ramachandran|first6=S.|last7=Cann|first7=H. M.|last8=Barsh|first8=G. S.|last9=Feldman|first9=M.|year=2008|title=Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation|journal=Science|volume=319|issue=5866|pages=1100–1104|bibcode=2008Sci...319.1100L|doi=10.1126/science.1153717|pmid=18292342|last10=Cavalli-Sforza|first10=L. L.|last11=Myers|first11=R. M.|s2cid=53541133}}</ref><ref>{{Cite journal|last1=Jakobsson|first1=M.|last2=Scholz|first2=S. W.|last3=Scheet|first3=P.|last4=Gibbs|first4=J. R.|last5=Vanliere|first5=J. M.|last6=Fung|first6=H. C.|last7=Szpiech|first7=Z. A.|last8=Degnan|first8=J. H.|last9=Wang|first9=K.|year=2008|title=Genotype, haplotype and copy-number variation in worldwide human populations|journal=Nature|volume=451|issue=7181|pages=998–1003|bibcode=2008Natur.451..998J|doi=10.1038/nature06742|pmid=18288195|last10=Guerreiro|first10=R.|last11=Bras|first11=J. M.|last12=Schymick|first12=J. C.|last13=Hernandez|first13=D. G.|last14=Traynor|first14=B. J.|last15=Simon-Sanchez|first15=J.|last16=Matarin|first16=M.|last17=Britton|first17=A.|last18=Van De Leemput|first18=J.|last19=Rafferty|first19=I.|last20=Bucan|first20=M.|last21=Cann|first21=H. M.|last22=Hardy|first22=J. A.|last23=Rosenberg|first23=N. A.|last24=Singleton|first24=A. B.|hdl=2027.42/62552|s2cid=11074384|url=https://deepblue.lib.umich.edu/bitstream/2027.42/62552/1/nature06742.pdf|hdl-access=free}}</ref><ref>{{Cite journal|last1=Xing|first1=J.|last2=Watkins|first2=W. S.|last3=Witherspoon|first3=D. J.|last4=Zhang|first4=Y.|last5=Guthery|first5=S. L.|last6=Thara|first6=R.|last7=Mowry|first7=B. J.|last8=Bulayeva|first8=K.|last9=Weiss|first9=R. B.|year=2009|title=Fine-scaled human genetic structure revealed by SNP microarrays|journal=Genome Research|volume=19|issue=5|pages=815–825|doi=10.1101/gr.085589.108|pmc=2675970|pmid=19411602|last10=Jorde|first10=L. B.}}</ref><ref>{{Cite journal|last1=López Herráez|first1=D.|last2=Bauchet|first2=M.|last3=Tang|first3=K.|last4=Theunert|first4=C.|last5=Pugach|first5=I.|last6=Li|first6=J.|last7=Nandineni|first7=M. R.|last8=Gross|first8=A.|last9=Scholz|first9=M.|year=2009|editor1-last=Hawks|editor1-first=John|title=Genetic Variation and Recent Positive Selection in Worldwide Human Populations: Evidence from Nearly 1 Million SNPs|journal=PLOS ONE|volume=4|issue=11|pages=e7888|bibcode=2009PLoSO...4.7888L|doi=10.1371/journal.pone.0007888|pmc=2775638|pmid=19924308|last10=Stoneking|first10=M.|doi-access=free}}</ref> |
||
Focus on study of structure has been criticized for giving the general public a misleading impression of human genetic variation, obscuring the general finding that genetic variants which are limited to one region tend to be rare within that region, variants that are common within a region tend to be shared across the globe, and most differences between individuals, whether they come from the same region or different regions, are due to global variants.<ref name="pmid33350384">{{cite journal |
Focus on study of structure has been criticized for giving the general public a misleading impression of human genetic variation, obscuring the general finding that genetic variants which are limited to one region tend to be rare within that region, variants that are common within a region tend to be shared across the globe, and most differences between individuals, whether they come from the same region or different regions, are due to global variants.<ref name="pmid33350384">{{cite journal |vauthors=Biddanda A, Rice DP, Novembre J| title=A variant-centric perspective on geographic patterns of human allele frequency variation. | journal=eLife | year= 2020 | volume= 9 | pmid=33350384 | doi=10.7554/eLife.60107 | pmc=7755386 | doi-access=free }}</ref> |
||
==== {{anchor|Genetic distance}}Distance ==== |
==== {{anchor|Genetic distance}}Distance ==== |
||
| Line 77: | Line 92: | ||
| issue = 2 |
| issue = 2 |
||
| pages = 141–147 |
| pages = 141–147 |
||
| last = Harpending |
| last = Harpending |
||
| first = Henry |
|||
| title = Kinship and Population Subdivision |
| title = Kinship and Population Subdivision |
||
| journal = [[Population & Environment]] |
| journal = [[Population & Environment]] |
||
| Line 84: | Line 100: | ||
| s2cid = 15208802 |
| s2cid = 15208802 |
||
| url = http://www.humanbiologicaldiversity.com/articles/Harpending,%20Henry.%20%22Kinship%20and%20Population%20Subdivision.%22%20Population%20and%20Environment%2024,%20no.%202%20(2002).pdf |
| url = http://www.humanbiologicaldiversity.com/articles/Harpending,%20Henry.%20%22Kinship%20and%20Population%20Subdivision.%22%20Population%20and%20Environment%2024,%20no.%202%20(2002).pdf |
||
| access-date = 2017-08-22 |
|||
| archive-date = 2017-06-28 |
|||
| archive-url = https://web.archive.org/web/20170628140206/http://www.humanbiologicaldiversity.com/articles/Harpending%2c%20Henry.%20%22Kinship%20and%20Population%20Subdivision.%22%20Population%20and%20Environment%2024%2c%20no.%202%20%282002%29.pdf |
|||
| url-status = live |
|||
}}</ref> |
}}</ref> |
||
==== Critiques of F<sub>ST</sub> ==== |
==== Critiques of F<sub>ST</sub> ==== |
||
While acknowledging that F<sub>ST</sub> remains useful, a number of scientists have written about other approaches to characterizing human genetic variation.<ref name="long">{{cite journal|author1=Long, J. C.|author2=Kittles, R. A.|year=2009|title=Human genetic diversity and the nonexistence of biological races|journal=Human Biology|volume=81|issue=5/6|pages=777–798|doi=10.3378/027.081.0621|pmid=20504196|name-list-style=amp|s2cid=30709062}}</ref><ref name="mountain">{{cite journal|author1=Mountain, J. L.|author2=Risch, N.|year=2004|title=Assessing genetic contributions to phenotypic differences among 'racial' and 'ethnic' groups|journal=Nature Genetics|volume=36|issue=11 Suppl|pages=S48–S53|doi=10.1038/ng1456|pmid=15508003|doi-access=free|name-list-style=amp}}</ref><ref name="pearse">{{cite journal| |
While acknowledging that F<sub>ST</sub> remains useful, a number of scientists have written about other approaches to characterizing human genetic variation.<ref name="long">{{cite journal|author1=Long, J. C.|author2=Kittles, R. A.|year=2009|title=Human genetic diversity and the nonexistence of biological races|journal=Human Biology|volume=81|issue=5/6|pages=777–798|doi=10.3378/027.081.0621|pmid=20504196|name-list-style=amp|s2cid=30709062}}</ref><ref name="mountain">{{cite journal|author1=Mountain, J. L.|author2=Risch, N.|year=2004|title=Assessing genetic contributions to phenotypic differences among 'racial' and 'ethnic' groups|journal=Nature Genetics|volume=36|issue=11 Suppl|pages=S48–S53|doi=10.1038/ng1456|pmid=15508003|doi-access=free|name-list-style=amp}}</ref><ref name="pearse">{{cite journal|last1=Pearse |first1=D. E.|last2=Crandall |first2=K. A.|author-link2=Keith A. Crandall|year=2004|title=Beyond FST: analysis of population genetic data for conservation|journal=Conservation Genetics|volume=5|issue=5|pages=585–602|doi=10.1007/s10592-003-1863-4|bibcode=2004ConG....5..585P |s2cid=22068080}}</ref> Long & Kittles (2009) stated that F<sub>ST</sub> failed to identify important variation and that when the analysis includes only humans, F<sub>ST</sub> = 0.119, but adding chimpanzees increases it only to F<sub>ST</sub> = 0.183.<ref name="long" /> Mountain & Risch (2004) argued that an F<sub>ST</sub> estimate of 0.10–0.15 does not rule out a genetic basis for phenotypic differences between groups and that a low F<sub>ST</sub> estimate implies little about the degree to which genes contribute to between-group differences.<ref name="mountain" /> Pearse & Crandall 2004 wrote that F<sub>ST</sub> figures cannot distinguish between a situation of high migration between populations with a long divergence time, and one of a relatively recent shared history but no ongoing gene flow.<ref name="pearse" /> In their 2015 article, Keith Hunley, Graciela Cabana, and Jeffrey Long (who had previously criticized Lewontin's statistical methodology with Rick Kittles<ref name=":0" />) recalculate the apportionment of human diversity using a more complex model than Lewontin and his successors. They conclude: "In sum, we concur with Lewontin's conclusion that Western-based racial classifications have no taxonomic significance, and we hope that this research, which takes into account our current understanding of the structure of human diversity, places his seminal finding on firmer evolutionary footing."<ref name=":1">{{Cite journal|last1=Hunley|first1=Keith L.|last2=Cabana|first2=Graciela S.|last3=Long|first3=Jeffrey C.|date=2015-12-01|title=The apportionment of human diversity revisited|journal=American Journal of Physical Anthropology|volume=160|issue=4|pages=561–569|doi=10.1002/ajpa.22899|issn=1096-8644|pmid=26619959|doi-access=free}}</ref> |
||
Anthropologists (such as [[C. Loring Brace]]),<ref>{{cite book|last=Brace|first=C. Loring|title="Race" is a Four-letter Word: The Genesis of the Concept|date=2005|publisher=Oxford University Press|isbn=978-0-19-517351-2|location=Oxford |
Anthropologists (such as [[C. Loring Brace]]),<ref>{{cite book|last=Brace|first=C. Loring|title="Race" is a Four-letter Word: The Genesis of the Concept|date=2005|publisher=Oxford University Press|isbn=978-0-19-517351-2|location=Oxford}}</ref> philosopher Jonathan Kaplan and geneticist [[Joseph L. Graves|Joseph Graves]]<ref>{{cite book|last=Graves|first=Joseph L|url=https://archive.org/details/emperorsnewcloth00grav|title=The Emperor's New Clothes: Biological Theories of Race at the Millennium|publisher=Rutgers University Press|year=2001|isbn=9780813528472|url-access=registration}}</ref> have argued that while it is possible to find biological and genetic variation roughly corresponding to race, this is true for almost all geographically distinct populations: the cluster structure of genetic data is dependent on the initial hypotheses of the researcher and the populations sampled. When one samples continental groups, the clusters become continental; with other sampling patterns, the clusters would be different. Weiss and Fullerton note that if one sampled only Icelanders, Mayans and Maoris, three distinct clusters would form; all other populations would be composed of [[genetic admixture]]s of Maori, Icelandic and Mayan material.<ref>{{cite journal|last1=Weiss|first1=Kenneth M.|last2=Fullerton|first2=Stephanie M.|year=2005|title=Racing around, getting nowhere|journal=Evolutionary Anthropology: Issues, News, and Reviews|volume=14|issue=5|pages=165–169|doi=10.1002/evan.20079|issn=1060-1538|s2cid=84927946}}</ref> Kaplan therefore concludes that, while differences in particular allele frequencies can be used to identify populations that loosely correspond to the racial categories common in Western social discourse, the differences are of no more biological significance than the differences found between any human populations (e.g., the Spanish and Portuguese).<ref>{{cite encyclopedia|title='Race': What Biology Can Tell Us about a Social Construct|encyclopedia=Encyclopedia of Life Sciences (ELS)|url=http://www.els.net/WileyCDA/ElsArticle/refId-a0005857.html|access-date=23 September 2014|date=17 January 2011|doi=10.1002/9780470015902.a0005857|isbn=978-0470016176|last1=Kaplan|first1=Jonathan Michael}}</ref> |
||
==={{anchor|Historic and geographic analysis of ancestry}}Historical and geographical analyses=== |
==={{anchor|Historic and geographic analysis of ancestry}}Historical and geographical analyses=== |
||
Current-population genetic structure does not imply that differing clusters or components indicate only one ancestral home per group; for example, a genetic cluster in the US comprises Hispanics with European, Native American and African ancestry.<ref name="Tang2005" /> |
Current-population genetic structure does not imply that differing clusters or components indicate only one ancestral home per group; for example, a genetic cluster in the US comprises Hispanics with European, Native American and African ancestry.<ref name="Tang2005" /> |
||
Geographic analyses attempt to identify places of origin, their relative importance and possible causes of genetic variation in an area. The results can be presented as maps showing genetic variation. Cavalli-Sforza and colleagues argue that if genetic variations are investigated, they often correspond to population migrations due to new sources of food, improved transportation or shifts in political power. For example, in Europe the most significant direction of genetic variation corresponds to the spread of agriculture from the Middle East to Europe between 10,000 and 6,000 years ago.<ref name="Cavalli-Sforza1997">Luigi Luca Cavalli-Sforza |
Geographic analyses attempt to identify places of origin, their relative importance and possible causes of genetic variation in an area. The results can be presented as maps showing genetic variation. Cavalli-Sforza and colleagues argue that if genetic variations are investigated, they often correspond to population migrations due to new sources of food, improved transportation or shifts in political power. For example, in Europe the most significant direction of genetic variation corresponds to the spread of agriculture from the Middle East to Europe between 10,000 and 6,000 years ago.<ref name="Cavalli-Sforza1997">{{cite journal |first=Luigi Luca |last=Cavalli-Sforza |title=Genes, peoples, and languages |journal=Proceedings of the National Academy of Sciences |year=1997 |volume=94 |issue=15 |pages=7719–7724 |doi=10.1073/pnas.94.15.7719|doi-access=free |bibcode=1997PNAS...94.7719C |pmc=33682 }}</ref> Such geographic analysis works best in the absence of recent large-scale, rapid migrations. |
||
Historic analyses use differences in genetic variation (measured by genetic distance) as a [[molecular clock]] indicating the evolutionary relation of species or groups, and can be used to create [[evolutionary tree]]s reconstructing population separations.<ref name="Cavalli-Sforza1997"/> |
Historic analyses use differences in genetic variation (measured by genetic distance) as a [[molecular clock]] indicating the evolutionary relation of species or groups, and can be used to create [[evolutionary tree]]s reconstructing population separations.<ref name="Cavalli-Sforza1997"/> |
||
| Line 101: | Line 121: | ||
==={{anchor|Self-identified race/ethnic group}}Self-identification studies=== |
==={{anchor|Self-identified race/ethnic group}}Self-identification studies=== |
||
Jorde and Wooding found that while clusters from genetic markers were correlated with some traditional concepts of race, the correlations were imperfect and imprecise due to the continuous and overlapping nature of genetic variation, noting that ancestry, which can be accurately determined, is not equivalent to the concept of race.<ref name= |
Jorde and Wooding found that while clusters from genetic markers were correlated with some traditional concepts of race, the correlations were imperfect and imprecise due to the continuous and overlapping nature of genetic variation, noting that ancestry, which can be accurately determined, is not equivalent to the concept of race.<ref name="Jorde2004"/> |
||
A 2005 study by Tang and colleagues used 326 genetic markers to determine genetic clusters. The 3,636 subjects, from the [[United States]] and [[Taiwan]], self-identified as belonging to white, African American, East Asian or Hispanic ethnic groups. The study found "nearly perfect correspondence between genetic cluster and SIRE for major ethnic groups living in the United States, with a discrepancy rate of only 0.14 percent".<ref name=Tang2005/> Paschou et al. found "essentially perfect" agreement between 51 self-identified populations of origin and the population's genetic structure, using 650,000 genetic markers. Selecting for informative genetic markers allowed a reduction to less than 650, while retaining near-total accuracy.<ref>{{cite journal | last1 = Paschou | first1 = Peristera | last2 = Lewis | first2 = Jamey | last3 = Javed | first3 = Asif | last4 = Drineas | first4 = Petros | year = 2010| title = Ancestry informative markers for fine-scale individual assignment to worldwide populations | url = https://hal.archives-ouvertes.fr/hal-00573484/document| journal = J Med Genet | volume = |
A 2005 study by Tang and colleagues used 326 genetic markers to determine genetic clusters. The 3,636 subjects, from the [[United States]] and [[Taiwan]], self-identified as belonging to white, African American, East Asian or Hispanic ethnic groups. The study found "nearly perfect correspondence between genetic cluster and SIRE for major ethnic groups living in the United States, with a discrepancy rate of only 0.14 percent".<ref name=Tang2005/> Paschou et al. found "essentially perfect" agreement between 51 self-identified populations of origin and the population's genetic structure, using 650,000 genetic markers. Selecting for informative genetic markers allowed a reduction to less than 650, while retaining near-total accuracy.<ref>{{cite journal | last1 = Paschou | first1 = Peristera | last2 = Lewis | first2 = Jamey | last3 = Javed | first3 = Asif | last4 = Drineas | first4 = Petros | year = 2010 | title = Ancestry informative markers for fine-scale individual assignment to worldwide populations | url = https://hal.archives-ouvertes.fr/hal-00573484/document | journal = J Med Genet | volume = 47 | issue = 12 | pages = 835–847 | doi = 10.1136/jmg.2010.078212 | pmid = 20921023 | s2cid = 6432430 | type = <!-- Submitted manuscript --> | access-date = 2018-11-04 | archive-date = 2018-11-05 | archive-url = https://web.archive.org/web/20181105064859/https://hal.archives-ouvertes.fr/hal-00573484/document | url-status = live }}</ref> |
||
Correspondence between genetic clusters in a population (such as the current US population) and self-identified race or ethnic groups does not mean that such a cluster (or group) corresponds to only one ethnic group. African Americans have an estimated 20–25-percent European genetic admixture; Hispanics have European, Native American and African ancestry.<ref name=Tang2005/> In Brazil there has been extensive admixture between Europeans, Amerindians and Africans. As a result, skin color differences within the population are not gradual, and there are relatively weak associations between self-reported race and African ancestry.<ref>{{Cite journal | last1 = Pena | first1 = Sérgio D. J.| last2 = Di Pietro | first2 = Giuliano| last3 = Fuchshuber-Moraes | first3 = Mateus| last4 = Genro | first4 = Julia Pasqualini| last5 = Hutz | first5 = Mara H.| last6 = Kehdy | first6 = Fernanda de Souza Gomes| last7 = Kohlrausch | first7 = Fabiana| last8 = Magno | first8 = Luiz Alexandre Viana| last9 = Montenegro | first9 = Raquel Carvalho| last10 = Moraes | first10 = Manoel Odorico| last11 = de Moraes | first11 = Maria Elisabete Amaral| last12 = de Moraes | first12 = Milene Raiol| last13 = Ojopi | first13 = Élida B.| last14 = Perini | first14 = Jamila A.| last15 = Racciopi | first15 = Clarice| last16 = Ribeiro-dos-Santos | first16 = Ândrea Kely Campos| last17 = Rios-Santos | first17 = Fabrício| last18 = Romano-Silva | first18 = Marco A.| last19 = Sortica | first19 = Vinicius A.| last20 = Suarez-Kurtz | first20 = Guilherme| editor1-last = Harpending | editor1-first = Henry | title = The Genomic Ancestry of Individuals from Different Geographical Regions of Brazil is More Uniform Than Expected | doi = 10.1371/journal.pone.0017063 | journal = [[PLoS ONE]]| volume = 6 | issue = 2 | pages = e17063 | year = 2011 | pmid = 21359226| pmc = 3040205| bibcode = 2011PLoSO...617063P| doi-access = free}}</ref><ref>{{Cite journal | last1 = Parra | first1 = F. C. | doi = 10.1073/pnas.0126614100 | title = Color and genomic ancestry in Brazilians | journal = Proceedings of the National Academy of Sciences | volume = 100 | pages = 177–182 | year = 2002 | pmid = |
Correspondence between genetic clusters in a population (such as the current US population) and self-identified race or ethnic groups does not mean that such a cluster (or group) corresponds to only one ethnic group. African Americans have an estimated 20–25-percent European genetic admixture; Hispanics have European, Native American and African ancestry.<ref name=Tang2005/> In Brazil there has been extensive admixture between Europeans, Amerindians and Africans. As a result, skin color differences within the population are not gradual, and there are relatively weak associations between self-reported race and African ancestry.<ref>{{Cite journal | last1 = Pena | first1 = Sérgio D. J.| last2 = Di Pietro | first2 = Giuliano| last3 = Fuchshuber-Moraes | first3 = Mateus| last4 = Genro | first4 = Julia Pasqualini| last5 = Hutz | first5 = Mara H.| last6 = Kehdy | first6 = Fernanda de Souza Gomes| last7 = Kohlrausch | first7 = Fabiana| last8 = Magno | first8 = Luiz Alexandre Viana| last9 = Montenegro | first9 = Raquel Carvalho| last10 = Moraes | first10 = Manoel Odorico| last11 = de Moraes | first11 = Maria Elisabete Amaral| last12 = de Moraes | first12 = Milene Raiol| last13 = Ojopi | first13 = Élida B.| last14 = Perini | first14 = Jamila A.| last15 = Racciopi | first15 = Clarice| last16 = Ribeiro-dos-Santos | first16 = Ândrea Kely Campos| last17 = Rios-Santos | first17 = Fabrício| last18 = Romano-Silva | first18 = Marco A.| last19 = Sortica | first19 = Vinicius A.| last20 = Suarez-Kurtz | first20 = Guilherme| editor1-last = Harpending | editor1-first = Henry | title = The Genomic Ancestry of Individuals from Different Geographical Regions of Brazil is More Uniform Than Expected | doi = 10.1371/journal.pone.0017063 | journal = [[PLoS ONE]]| volume = 6 | issue = 2 | pages = e17063 | year = 2011 | pmid = 21359226| pmc = 3040205| bibcode = 2011PLoSO...617063P| doi-access = free}}</ref><ref>{{Cite journal | last1 = Parra | first1 = F. C. | doi = 10.1073/pnas.0126614100 | title = Color and genomic ancestry in Brazilians | journal = Proceedings of the National Academy of Sciences | volume = 100 | pages = 177–182 | year = 2002 | pmid = 12509516| pmc = 140919|bibcode = 2003PNAS..100..177P| issue=1| doi-access = free }}</ref> Ethnoracial self- classification in Brazilians is certainly not random with respect to genome individual ancestry, but the strength of the association between the phenotype and median proportion of African ancestry varies largely across population.<ref>{{Cite journal | doi=10.1038/srep09812| pmid=25913126| pmc=5386196| title=Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative)| journal=Scientific Reports| volume=5| pages=9812| year=2015| last1=Lima-Costa| first1=M. Fernanda| last2=Rodrigues| first2=Laura C.| last3=Barreto| first3=Maurício L.| last4=Gouveia| first4=Mateus| last5=Horta| first5=Bernardo L.| last6=Mambrini| first6=Juliana| last7=Kehdy| first7=Fernanda S. G.| last8=Pereira| first8=Alexandre| last9=Rodrigues-Soares| first9=Fernanda| last10=Victora| first10=Cesar G.| last11=Tarazona-Santos| first11=Eduardo| last12=Cesar| first12=Cibele C.| last13=Conceição| first13=Jackson S.| last14=Costa| first14=Gustavo N. O.| last15=Esteban| first15=Nubia| last16=Fiaccone| first16=Rosemeire L.| last17=Figueiredo| first17=Camila A.| last18=Firmo| first18=Josélia O. A.| last19=Horimoto| first19=Andrea R. V. R.| last20=Leal| first20=Thiago P.| last21=Machado| first21=Moara| last22=Magalhães| first22=Wagner C. S.| last23=De Oliveira| first23=Isabel Oliveira| last24=Peixoto| first24=Sérgio V.| last25=Rodrigues| first25=Maíra R.| last26=Santos| first26=Hadassa C.| last27=Silva| first27=Thiago M.| bibcode=2015NatSR...5E9812.}}</ref> |
||
=== {{anchor|Continuous or discontinuous increase in genetic distance}}Critique of genetic-distance studies and clusters === |
=== {{anchor|Continuous or discontinuous increase in genetic distance}}Critique of genetic-distance studies and clusters === |
||
[[File:IBD SIM.png|thumb|upright|alt=Colored circles, illustrating gene-pool changes|A change in a [[gene pool]] may be abrupt or [[Cline (biology)|clinal]].]] Genetic distances generally increase continually with geographic distance, which makes a dividing line arbitrary. Any two neighboring settlements will exhibit some genetic difference from each other, which could be defined as a race. Therefore, attempts to classify races impose an artificial discontinuity on a naturally occurring phenomenon. This explains why studies on population genetic structure yield varying results, depending on methodology.<ref>Reanne Frank |
[[File:IBD SIM.png|thumb|upright|alt=Colored circles, illustrating gene-pool changes|A change in a [[gene pool]] may be abrupt or [[Cline (biology)|clinal]].]] Genetic distances generally increase continually with geographic distance, which makes a dividing line arbitrary. Any two neighboring settlements will exhibit some genetic difference from each other, which could be defined as a race. Therefore, attempts to classify races impose an artificial discontinuity on a naturally occurring phenomenon. This explains why studies on population genetic structure yield varying results, depending on methodology.<ref>{{cite web |first=Reanne |last=Frank |url=http://paa2006.princeton.edu/download.aspx?submissionId=61713 |title=Back with a Vengeance: the Reemergence of a Biological Conceptualization of Race in Research on Race/Ethnic Disparities in Health |archive-url=https://web.archive.org/web/20081201101017/http://paa2006.princeton.edu/download.aspx?submissionId=61713 |archive-date=2008-12-01 }}</ref> |
||
Rosenberg and colleagues (2005) have argued, based on cluster analysis of the 52 populations in the Human Genetic Diversity Panel, that populations do not always vary continuously and a population's genetic structure is consistent if enough genetic markers (and subjects) are included. |
Rosenberg and colleagues (2005) have argued, based on cluster analysis of the 52 populations in the Human Genetic Diversity Panel, that populations do not always vary continuously and a population's genetic structure is consistent if enough genetic markers (and subjects) are included. |
||
| Line 119: | Line 139: | ||
This applies to populations in their ancestral homes when migrations and gene flow were slow; large, rapid migrations exhibit different characteristics. Tang and colleagues (2004) wrote, "we detected only modest genetic differentiation between different current geographic locales within each race/ethnicity group. Thus, ancient geographic ancestry, which is highly correlated with self-identified race/ethnicity—as opposed to current residence—is the major determinant of genetic structure in the U.S. population".<ref name="Tang2005" />[[File:Rosenberg2007.png|thumb|right|150px|Gene clusters from Rosenberg (2006) for K=7 clusters. ([[Cluster analysis]] divides a dataset into any prespecified number of clusters.) Individuals have genes from multiple clusters. The cluster prevalent only among the [[Kalash people]] (yellow) only splits off at K=7 and greater.]] |
This applies to populations in their ancestral homes when migrations and gene flow were slow; large, rapid migrations exhibit different characteristics. Tang and colleagues (2004) wrote, "we detected only modest genetic differentiation between different current geographic locales within each race/ethnicity group. Thus, ancient geographic ancestry, which is highly correlated with self-identified race/ethnicity—as opposed to current residence—is the major determinant of genetic structure in the U.S. population".<ref name="Tang2005" />[[File:Rosenberg2007.png|thumb|right|150px|Gene clusters from Rosenberg (2006) for K=7 clusters. ([[Cluster analysis]] divides a dataset into any prespecified number of clusters.) Individuals have genes from multiple clusters. The cluster prevalent only among the [[Kalash people]] (yellow) only splits off at K=7 and greater.]] |
||
Cluster analysis has been criticized because the number of clusters to search for is decided in advance, with different values possible (although with varying degrees of probability).<ref>{{Cite book| publisher = Rutgers University Press| isbn = 978-0-8135-4324-6| editor1-last = Koenig| editor1-first = Barbara A.| editor2-last = Richardson| editor2-first = Sarah S.| editor3-last = Lee| editor3-first = Sandra Soo-Jin| last = Bolnick| first = Deborah A.| title = Revisiting race in a genomic age| chapter = Individual Ancestry Inference and the Reification of Race as a Biological Phenomenon| year = 2008}}</ref> [[Principal component analysis]] does not decide in advance how many components for which to search.<ref name=Patterson2006>{{Cite journal | last1 = Patterson | first1 = Nick | last2 = Price | first2 = Alkes L. | last3 = Reich | first3 = David | doi = 10.1371/journal.pgen.0020190 | title = Population Structure and Eigenanalysis | journal = PLOS Genetics | volume = 2 | issue = 12 | pages = e190 | year = 2006 | pmid = |
Cluster analysis has been criticized because the number of clusters to search for is decided in advance, with different values possible (although with varying degrees of probability).<ref>{{Cite book| publisher = Rutgers University Press| isbn = 978-0-8135-4324-6| editor1-last = Koenig| editor1-first = Barbara A.| editor2-last = Richardson| editor2-first = Sarah S.| editor-link2=Sarah S. Richardson|editor3-last = Lee| editor3-first = Sandra Soo-Jin| last = Bolnick| first = Deborah A.| title = Revisiting race in a genomic age| chapter = Individual Ancestry Inference and the Reification of Race as a Biological Phenomenon| year = 2008}}</ref> [[Principal component analysis]] does not decide in advance how many components for which to search.<ref name=Patterson2006>{{Cite journal | last1 = Patterson | first1 = Nick | last2 = Price | first2 = Alkes L. | last3 = Reich | first3 = David | doi = 10.1371/journal.pgen.0020190 | title = Population Structure and Eigenanalysis | journal = PLOS Genetics | volume = 2 | issue = 12 | pages = e190 | year = 2006 | pmid = 17194218| pmc =1713260 | doi-access = free }}</ref> |
||
| ⚫ | The 2002 study by [[Noah Rosenberg|Rosenberg]] et al. |
||
However, in addition to the five main supposedly geographical clusters, a sixth group, the [[Kalash people|Kalash]], a minority ethnic group in Pakistan, began to appear starting at K=6. The racial naturalist [[Nicholas Wade]] considers that the results "make no genetic or geographic sense". They are therefore omitted in his book [[A Troublesome Inheritance]] in favour of the K=5 cluster analysis. |
|||
| ⚫ | The 2002 study by [[Noah Rosenberg|Rosenberg]] et al. exemplifies why meanings of these clusterings can be disputable, though the study shows that at the K=5 cluster analysis, genetic clusterings roughly map onto each of the five major geographical regions.<ref name='Rosenberg2002'/> Similar results were gathered in further studies in 2005.<ref name="rosenberg2005">{{cite journal | last1 = Rosenberg | first1 = N. A. | author-link5 = Jonathan K. Pritchard | last2 = Mahajan | first2 = S. | last3 = Ramachandran | first3 = S. | last4 = Zhao | first4 = C. | last5 = Pritchard | first5 = J. K. | display-authors = etal | year = 2005 | title = Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure | journal = PLOS Genet | volume = 1 | issue = 6| page = e70 | doi = 10.1371/journal.pgen.0010070 | pmid = 16355252 | pmc=1310579 | doi-access = free }}</ref> |
||
This bias, however, is reflective of how the research is inherently flawed. The sample population is chosen with geographical representation and folk concepts of race in mind, instead of accounting for the genetic diversity within the different geographical regions. The Kalash did not fit into the general pattern for it had been a genetically isolated population that happened to be reflected in this study. Potentially numerous genetically drifted groups, such as the [[Uncontacted peoples|uncontacted]] [[Sentinelese]], are not represented in the study.{{citation needed|date=June 2021}} |
|||
=== Critique of ancestry-informative markers === |
=== Critique of ancestry-informative markers === |
||
[[Ancestry-informative marker]]s (AIMs) are a genealogy tracing technology that has come under much criticism due to its reliance on reference populations. In a 2015 article, Troy Duster outlines how contemporary technology allows the tracing of ancestral lineage but along only the lines of one maternal and one paternal line. That is, of 64 total great-great-great-great-grandparents, only one from each parent is identified, implying the other 62 ancestors are ignored in tracing efforts.<ref name="Duster 1–27">{{Cite journal|last=Duster|first=Troy|date=March 2015|title=A post-genomic surprise. The molecular reinscription of race in science, law and medicine|journal=The British Journal of Sociology|volume=66|issue=1|pages=1–27|doi=10.1111/1468-4446.12118|pmid=25789799|issn=0007-1315}}</ref> Furthermore, the 'reference populations' used as markers for membership of a particular group are designated arbitrarily and contemporarily. In other words, using populations who currently reside in given places as references for certain races and ethnic groups is unreliable due to the demographic changes which have occurred over many centuries in those places. Furthermore, ancestry-informative markers being widely shared among the whole human population, it is their frequency which is tested, not their mere absence/presence. A threshold of relative frequency has, therefore, to be set. According to Duster the criteria for setting such thresholds are a trade secret of the companies marketing the tests. Thus, we cannot say anything conclusive on whether they are appropriate. |
[[Ancestry-informative marker]]s (AIMs) are a genealogy tracing technology that has come under much criticism due to its reliance on reference populations. In a 2015 article, Troy Duster outlines how contemporary technology allows the tracing of ancestral lineage but along only the lines of one maternal and one paternal line. That is, of 64 total great-great-great-great-grandparents, only one from each parent is identified, implying the other 62 ancestors are ignored in tracing efforts.<ref name="Duster 1–27">{{Cite journal|last=Duster|first=Troy|date=March 2015|title=A post-genomic surprise. The molecular reinscription of race in science, law and medicine|journal=The British Journal of Sociology|volume=66|issue=1|pages=1–27|doi=10.1111/1468-4446.12118|pmid=25789799|issn=0007-1315}}</ref> Furthermore, the 'reference populations' used as markers for membership of a particular group are designated arbitrarily and contemporarily. In other words, using populations who currently reside in given places as references for certain races and ethnic groups is unreliable due to the demographic changes which have occurred over many centuries in those places. Furthermore, ancestry-informative markers being widely shared among the whole human population, it is their frequency which is tested, not their mere absence/presence. A threshold of relative frequency has, therefore, to be set. According to Duster, the criteria for setting such thresholds are a trade secret of the companies marketing the tests. Thus, we cannot say anything conclusive on whether they are appropriate. |
||
Results of AIMs are extremely sensitive to where this bar is set.<ref>Fullwiley, D. (2008). "The Biologistical Construction of Race: 'Admixture' Technology and the New Genetic Medicine". ''Social Studies of Science'', 38(5), 695–735. {{doi|10.1177/0306312708090796}} |
Results of AIMs are extremely sensitive to where this bar is set.<ref>Fullwiley, D. (2008). "The Biologistical Construction of Race: 'Admixture' Technology and the New Genetic Medicine". ''Social Studies of Science'', 38(5), 695–735. {{doi|10.1177/0306312708090796}} |
||
</ref> Given that many genetic traits are found very similar amid many different populations, the |
</ref> Given that many genetic traits are found to be very similar amid many different populations, the designated threshold frequencies are very important. This can also lead to mistakes, given that many populations may share the same patterns, if not exactly the same genes. "This means that someone from Bulgaria whose ancestors go back to the fifteenth century could (and sometime does) map as partly 'Native American{{'"}}.<ref name="Duster 1–27"/> This happens because AIMs rely on a '100% purity' assumption of reference populations. That is, they assume that a pattern of traits would ideally be a necessary and sufficient condition for assigning an individual to an ancestral reference populations. |
||
==Race, genetics, and medicine== |
==Race, genetics, and medicine== |
||
{{Main|Race and health}} |
{{Main|Race and health}} |
||
There are certain statistical differences between racial groups in susceptibility to certain diseases.<ref name=Gitschier2005>{{cite journal |author=Risch N |title=The whole side of it--an interview with Neil Risch by Jane Gitschier |journal=PLOS Genetics |volume=1 |issue=1 |pages=e14 |date=July 2005 |pmid=17411332 |doi=10.1371/journal.pgen.0010014 |pmc=1183530}}</ref> Genes change in response to local diseases; for example, people who are [[Duffy antigen system|Duffy-negative]] tend to have a higher resistance to malaria. The Duffy negative phenotype is highly frequent in central Africa and the frequency decreases with distance away from Central Africa, with higher frequencies in global populations with high degrees of recent African immigration. This suggests that the Duffy negative genotype evolved in Sub-Saharan Africa and was subsequently positively selected for in the Malaria endemic zone.<ref> |
There are certain statistical differences between racial groups in susceptibility to certain diseases.<ref name=Gitschier2005>{{cite journal |author=Risch N |title=The whole side of it--an interview with Neil Risch by Jane Gitschier |journal=PLOS Genetics |volume=1 |issue=1 |pages=e14 |date=July 2005 |pmid=17411332 |doi=10.1371/journal.pgen.0010014 |pmc=1183530 |doi-access=free }}</ref> Genes change in response to local diseases; for example, people who are [[Duffy antigen system|Duffy-negative]] tend to have a higher resistance to malaria. The Duffy negative phenotype is highly frequent in central Africa and the frequency decreases with distance away from Central Africa, with higher frequencies in global populations with high degrees of recent African immigration. This suggests that the Duffy negative genotype evolved in Sub-Saharan Africa and was subsequently positively selected for in the Malaria endemic zone.<ref>{{cite web |url=http://sickle.bwh.harvard.edu/malaria_sickle.html |title=Malaria and the Red Cell |archive-url=https://web.archive.org/web/20111127201806/http://sickle.bwh.harvard.edu/malaria_sickle.html |archive-date=2011-11-27 |website=Harvard University |year=2002}}</ref> A number of genetic conditions prevalent in malaria-endemic areas may provide [[genetic resistance to malaria]], including [[sickle cell disease]], [[thalassaemias]] and [[glucose-6-phosphate dehydrogenase]]. [[Cystic fibrosis]] is the most common life-limiting [[autosomal recessive]] disease among people of European ancestry; a hypothesized [[heterozygote advantage]], providing resistance to diseases earlier common in Europe, has been challenged.<ref>{{cite journal |vauthors=Högenauer C, Santa Ana CA, Porter JL, et al |title=Active intestinal chloride secretion in human carriers of cystic fibrosis mutations: an evaluation of the hypothesis that heterozygotes have subnormal active intestinal chloride secretion |journal=Am. J. Hum. Genet. |volume=67 |issue=6 |pages=1422–1427 |date=December 2000 |pmid=11055897 |pmc=1287919 |doi=10.1086/316911 }}</ref> Scientists Michael Yudell, Dorothy Roberts, Rob DeSalle, and Sarah Tishkoff argue that using these associations in the practice of medicine has led doctors to overlook or misidentify disease: "For example, hemoglobinopathies can be misdiagnosed because of the identification of sickle-cell as a 'Black' disease and thalassemia as a 'Mediterranean' disease. Cystic fibrosis is underdiagnosed in populations of African ancestry, because it is thought of as a 'White' disease."<ref name="Yudell2016"/> |
||
Information about a person's population of origin may aid in [[diagnosis]], and adverse drug responses may vary by group.<ref name="Rosenberg2002"/>{{Dubious|date=September 2018}} Because of the correlation between self-identified race and genetic clusters, medical treatments influenced by genetics have varying rates of success between self-defined racial groups.<ref>{{cite journal | year = 2001 | title = Racial Differences in the Response to Drugs — Pointers to Genetic Differences | url = http://content.nejm.org/cgi/content/extract/344/18/1393 | journal = New England Journal of Medicine | volume = 344 | issue = 18| pages = 1393–1396 |doi=10.1056/NEJM200105033441810 | pmid = 11333999 | last1 = Schwartz | first1 = Robert S.}}</ref> For this reason, some physicians{{who|date=September 2014}} consider a patient's race in choosing the most effective treatment,<ref>{{cite journal | last1 = Bloche | first1 = Gregg M. | year = 2004 | title = Race-Based Therapeutics | journal = New England Journal of Medicine | volume = 351 | issue = 20| pages = 2035–2037 | doi=10.1056/nejmp048271 | pmid=15533852}}</ref> and some drugs are marketed with race-specific instructions.<ref>[http://www.emedicinehealth.com/drug-rosuvastatin/article_em.htm Drug information for the drug Crestor]. Warnings for this drug state, "People of Asian descent may absorb rosuvastatin at a higher rate than other people. Make sure your doctor knows if you are Asian. You may need a lower than normal starting dose."</ref> Jorde and Wooding (2004) have argued that because of genetic variation within racial groups, when "it finally becomes feasible and available, individual genetic assessment of relevant genes will probably prove more useful than race in medical decision making". However, race continues to be a factor when examining groups (such as epidemiologic research).<ref name= |
Information about a person's population of origin may aid in [[diagnosis]], and adverse drug responses may vary by group.<ref name="Rosenberg2002"/>{{Dubious|date=September 2018}} Because of the correlation between self-identified race and genetic clusters, medical treatments influenced by genetics have varying rates of success between self-defined racial groups.<ref>{{cite journal | year = 2001 | title = Racial Differences in the Response to Drugs — Pointers to Genetic Differences | url = http://content.nejm.org/cgi/content/extract/344/18/1393 | journal = New England Journal of Medicine | volume = 344 | issue = 18 | pages = 1393–1396 | doi = 10.1056/NEJM200105033441810 | pmid = 11333999 | last1 = Schwartz | first1 = Robert S. | access-date = 2009-10-28 | archive-date = 2003-09-01 | archive-url = https://web.archive.org/web/20030901034140/http://content.nejm.org/cgi/content/extract/344/18/1393 | url-status = dead }}</ref> For this reason, some physicians{{who|date=September 2014}} consider a patient's race in choosing the most effective treatment,<ref>{{cite journal | last1 = Bloche | first1 = Gregg M. | year = 2004 | title = Race-Based Therapeutics | journal = New England Journal of Medicine | volume = 351 | issue = 20| pages = 2035–2037 | doi=10.1056/nejmp048271 | pmid=15533852}}</ref> and some drugs are marketed with race-specific instructions.<ref>[http://www.emedicinehealth.com/drug-rosuvastatin/article_em.htm Drug information for the drug Crestor] {{Webarchive|url=https://web.archive.org/web/20090926084534/http://www.emedicinehealth.com/drug-rosuvastatin/article_em.htm |date=2009-09-26 }}. Warnings for this drug state, "People of Asian descent may absorb rosuvastatin at a higher rate than other people. Make sure your doctor knows if you are Asian. You may need a lower than normal starting dose."</ref> Jorde and Wooding (2004) have argued that because of genetic variation within racial groups, when "it finally becomes feasible and available, individual genetic assessment of relevant genes will probably prove more useful than race in medical decision making". However, race continues to be a factor when examining groups (such as epidemiologic research).<ref name="Jorde2004"/> Some doctors and scientists such as geneticist [[Neil Risch]] argue that using self-identified race as a proxy for ancestry is necessary to be able to get a sufficiently broad sample of different ancestral populations, and in turn to be able to provide health care that is tailored to the needs of minority groups.<ref>{{cite journal | last1 = Risch | first1 = N. | last2 = Burchard | first2 = E. | last3 = Ziv | first3 = E. | last4 = Tang | first4 = H. | year = 2002 | title = Categorization of humans in biomedical research: genes, race and disease | doi = 10.1186/gb-2002-3-7-comment2007 | journal = Genome Biol | volume = 3 | issue = 7| pages = 1–12 | pmid=12184798 | pmc=139378 | doi-access = free }}</ref> |
||
===Usage in scientific journals=== |
===Usage in scientific journals=== |
||
| Line 145: | Line 161: | ||
===Gene-environment interactions=== |
===Gene-environment interactions=== |
||
Lorusso and Bacchini<ref name= |
Lorusso and Bacchini<ref name='LorussoBacchini'/> argue that self-identified race is of greater use in medicine as it correlates strongly with risk-related [[exposome]]s that are potentially heritable when they become embodied in the [[epigenome]]. They summarise evidence of the link between racial discrimination and health outcomes due to poorer food quality, access to healthcare, housing conditions, education, access to information, exposure to infectious agents and toxic substances, and material scarcity. They also cite evidence that this process can work positively – for example, the psychological advantage of perceiving oneself at the top of a social hierarchy is linked to improved health. However they caution that the effects of discrimination do not offer a complete explanation for differential rates of disease and risk factors between racial groups, and the employment of self-identified race has the potential to reinforce racial inequalities. |
||
== Objections to racial naturalism == |
== Objections to racial naturalism == |
||
Racial naturalism is the view that racial classifications are grounded in objective patterns of genetic similarities and differences. Proponents of this view have justified it using the scientific evidence described above. However, this view is controversial and philosophers<ref>{{ |
Racial naturalism is the view that racial classifications are grounded in objective patterns of genetic similarities and differences. Proponents of this view have justified it using the scientific evidence described above. However, this view is controversial and philosophers<ref>{{cite book |chapter=Revelations |chapter-url=http://dx.doi.org/10.1057/9780230362987.0004 |title=Shut Up and Listen |year=2011 |publisher=Palgrave Macmillan |doi=10.1057/9780230362987.0004 |isbn=978-0-230-36298-7 |access-date=2021-01-28}}.</ref> of race have put forward four main objections to it. |
||
Semantic objections, such as the discreteness objection, argue that the human populations picked out in population-genetic research are not races and do not correspond to what "race" means in the United States. "The discreteness objection does not require there to be no genetic admixture in the human species in order for there to be US 'racial groups' ... rather ... what the objection claims is that membership in US racial groups is different from membership in continental populations. ... Thus, strictly speaking, Blacks are not identical to Africans, Whites are not identical to Eurasians, Asians are not identical to East Asians and so forth."<ref name=spencer>{{cite journal |last1=Spencer |first1=Quayshawn |title=Philosophy of race meets population genetics |journal=Studies in History and Philosophy of Biological and Biomedical Sciences |date=2015 |volume=52 |pages=49|doi=10.1016/j.shpsc.2015.04.003 |pmid=25963045 }}</ref> Therefore, it could be argued that scientific research is not really about race. |
Semantic objections, such as the discreteness objection, argue that the human populations picked out in population-genetic research are not races and do not correspond to what "race" means in the United States. "The discreteness objection does not require there to be no genetic admixture in the human species in order for there to be US 'racial groups' ... rather ... what the objection claims is that membership in US racial groups is different from membership in continental populations. ... Thus, strictly speaking, Blacks are not identical to Africans, Whites are not identical to Eurasians, Asians are not identical to East Asians and so forth."<ref name=spencer>{{cite journal |last1=Spencer |first1=Quayshawn |title=Philosophy of race meets population genetics |journal=Studies in History and Philosophy of Biological and Biomedical Sciences |date=2015 |volume=52 |pages=49|doi=10.1016/j.shpsc.2015.04.003 |pmid=25963045 }}</ref> Therefore, it could be argued that scientific research is not really about race. |
||
| Line 158: | Line 174: | ||
A different objection states that US racial groups are not biologically real because they are not objectively real in the sense of existing independently of some mental state of humans. Proponents of this second metaphysical objection include [[Naomi Zack]] and Ron Sundstrom.<ref name="zach" /><ref>{{cite journal |last1=Sundstrom |first1=R |date=2002 |title=Race as a human kind |journal=Philosophy & Social Criticism |volume=28 |pages=91–115|doi=10.1177/0191453702028001592 |s2cid=145381236 }}</ref> Spencer argues that an entity can be both biologically real and socially constructed. Spencer states that in order to accurately capture real biological entities, social factors must also be considered.{{citation needed|date=April 2021}}{{undue weight inline|date=April 2021}} |
A different objection states that US racial groups are not biologically real because they are not objectively real in the sense of existing independently of some mental state of humans. Proponents of this second metaphysical objection include [[Naomi Zack]] and Ron Sundstrom.<ref name="zach" /><ref>{{cite journal |last1=Sundstrom |first1=R |date=2002 |title=Race as a human kind |journal=Philosophy & Social Criticism |volume=28 |pages=91–115|doi=10.1177/0191453702028001592 |s2cid=145381236 }}</ref> Spencer argues that an entity can be both biologically real and socially constructed. Spencer states that in order to accurately capture real biological entities, social factors must also be considered.{{citation needed|date=April 2021}}{{undue weight inline|date=April 2021}} |
||
It has been argued that knowledge of a person's race is limited in value, since people of the same race vary from one another.<ref name=" |
It has been argued that knowledge of a person's race is limited in value, since people of the same race vary from one another.<ref name="Jorde2004"/> [[David J. Witherspoon]] and colleagues have argued that when individuals are assigned to population groups, two randomly chosen individuals from different populations can resemble each other more than a randomly chosen member of their own group. They found that many thousands of genetic markers had to be used for the answer to "How often is a pair of individuals from one population genetically more dissimilar than two individuals chosen from two different populations?" to be "Never". This assumed three population groups, separated by large geographic distances (European, African and East Asian). The global human population is more complex, and studying a large number of groups would require an increased number of markers for the same answer. They conclude that "caution should be used when using geographic or genetic ancestry to make inferences about individual phenotypes",<ref>{{Cite journal|vauthors=Witherspoon DJ, Wooding S, Rogers AR, etal|date=May 2007|title=Genetic Similarities Within and Between Human Populations|journal=Genetics|volume=176|issue=1|pages=351–9|doi=10.1534/genetics.106.067355|pmc=1893020|pmid=17339205}}</ref> and "The fact that, given enough genetic data, individuals can be correctly assigned to their populations of origin is compatible with the observation that most human genetic variation is found within populations, not between them. It is also compatible with our finding that, even when the most distinct populations are considered and hundreds of loci are used, individuals are frequently more similar to members of other populations than to members of their own population".<ref>{{Cite journal|vauthors=Witherspoon DJ, Wooding S, Rogers AR, etal|date=May 2007|title=Genetic Similarities Within and Between Human Populations|journal=Genetics|volume=176|issue=1|page=358|doi=10.1534/genetics.106.067355|pmc=1893020|pmid=17339205}}</ref> |
||
This is similar to the conclusion reached by anthropologist [[Norman Sauer]] in a 1992 article on the ability of forensic anthropologists to assign "race" to a skeleton, based on craniofacial features and limb morphology. Sauer said, "the successful assignment of race to a skeletal specimen is not a vindication of the race concept, but rather a prediction that an individual, while alive was assigned to a particular socially constructed 'racial' category. A specimen may display features that point to African ancestry. In this country that person is likely to have been labeled Black regardless of whether or not such a race actually exists in nature".<ref name="Sauer 1992">{{cite journal|last1=Sauer|first1=N. J.|date=January 1992|title=Forensic anthropology and the concept of race: if races don't exist, why are forensic anthropologists so good at identifying them?|journal=Social Science & Medicine|volume=34|issue=2|pages=107–111|doi=10.1016/0277-9536(92)90086-6|pmid=1738862}}</ref> |
This is similar to the conclusion reached by anthropologist [[Norman Sauer]] in a 1992 article on the ability of forensic anthropologists to assign "race" to a skeleton, based on craniofacial features and limb morphology. Sauer said, "the successful assignment of race to a skeletal specimen is not a vindication of the race concept, but rather a prediction that an individual, while alive was assigned to a particular socially constructed 'racial' category. A specimen may display features that point to African ancestry. In this country that person is likely to have been labeled Black regardless of whether or not such a race actually exists in nature".<ref name="Sauer 1992">{{cite journal|last1=Sauer|first1=N. J.|date=January 1992|title=Forensic anthropology and the concept of race: if races don't exist, why are forensic anthropologists so good at identifying them?|journal=Social Science & Medicine|volume=34|issue=2|pages=107–111|doi=10.1016/0277-9536(92)90086-6|pmid=1738862}}</ref> |
||
| Line 167: | Line 183: | ||
Duster elaborates by putting forward the example of [[Pima people|the Pima]] of [[Arizona]], a population suffering from disproportionately high rates of [[Diabetes mellitus|diabetes]]. The reason for such, he argues, was not necessarily a result of the prevalence of the [[FABP2]] gene, which is associated with [[insulin resistance]]. Rather he argues that scientists often discount the lifestyle implications under specific socio-historical contexts. For instance, near the end of the 19th century, the Pima economy was predominantly agriculture-based. However, as the European American population settles into traditionally Pima territory, the Pima lifestyles became heavily Westernised. Within three decades, the incidence of diabetes increased multiple folds. Governmental provision of free relatively high-fat food to alleviate the prevalence of poverty in the population is noted as an explanation of this phenomenon.<ref>{{Cite journal |last=Duster |first=Troy |date=2015 |title=A post-genomic surprise. The molecular reinscription of race in science, law and medicine |journal=The British Journal of Sociology |volume=66|issue=1 |pages=1–27 |doi=10.1111/1468-4446.12118 |pmid=25789799 }}</ref> |
Duster elaborates by putting forward the example of [[Pima people|the Pima]] of [[Arizona]], a population suffering from disproportionately high rates of [[Diabetes mellitus|diabetes]]. The reason for such, he argues, was not necessarily a result of the prevalence of the [[FABP2]] gene, which is associated with [[insulin resistance]]. Rather he argues that scientists often discount the lifestyle implications under specific socio-historical contexts. For instance, near the end of the 19th century, the Pima economy was predominantly agriculture-based. However, as the European American population settles into traditionally Pima territory, the Pima lifestyles became heavily Westernised. Within three decades, the incidence of diabetes increased multiple folds. Governmental provision of free relatively high-fat food to alleviate the prevalence of poverty in the population is noted as an explanation of this phenomenon.<ref>{{Cite journal |last=Duster |first=Troy |date=2015 |title=A post-genomic surprise. The molecular reinscription of race in science, law and medicine |journal=The British Journal of Sociology |volume=66|issue=1 |pages=1–27 |doi=10.1111/1468-4446.12118 |pmid=25789799 }}</ref> |
||
Lorusso and Bacchini argue against the assumption that "self-identified race is a good proxy for a specific genetic ancestry"<ref name= |
Lorusso and Bacchini argue against the assumption that "self-identified race is a good proxy for a specific genetic ancestry"<ref name='LorussoBacchini' /> on the basis that self-identified race is complex: it depends on a range of psychological, cultural and social factors, and is therefore "not a robust proxy for genetic ancestry".<ref name="ambiguous">{{Cite journal|first1=L. M. |last1=Hunt |first2=M.S. |last2=Megyesi|date=Fall 2007|title=The ambiguous meanings of the racial/ethnic categories routinely used in human genetics research|journal=Social Science and Medicine|volume=66|issue=2 |pages=349–361|via=Science Direct Assets|doi=10.1016/j.socscimed.2007.08.034|pmid=17959289|pmc=2213883}}</ref> Furthermore, they explain that an individual's self-identified race is made up of further, collectively arbitrary factors: personal opinions about what race is and the extent to which it should be taken into consideration in everyday life. Furthermore, individuals who share a genetic ancestry may differ in their racial self-identification across historical or socioeconomic contexts. From this, Lorusso and Bacchini conclude that the accuracy in the prediction of genetic ancestry on the basis of self-identification is low, specifically in racially admixed populations born out of complex ancestral histories. |
||
== See also == |
== See also == |
||
| Line 174: | Line 190: | ||
* {{annotated link|Human subspecies}} |
* {{annotated link|Human subspecies}} |
||
* {{annotated link|Human Genetic Diversity: Lewontin's Fallacy}} |
* {{annotated link|Human Genetic Diversity: Lewontin's Fallacy}} |
||
* [[Zionism, race and genetics]] |
|||
==References== |
==References== |
||
| Line 179: | Line 196: | ||
== Further reading == |
== Further reading == |
||
*{{cite book |last=Glasgow |first=Joshua |year=2009 |title=[[A Theory of Race]] |location=New York |publisher=Routledge |isbn=9780415990721}} |
|||
*{{cite journal |vauthors=Helms JE, Jernigan M, Mascher J |title=The meaning of race in psychology and how to change it: a methodological perspective |journal=The American Psychologist |volume=60 |issue=1 |pages=27–36 |date=January 2005 |pmid=15641919 |doi=10.1037/0003-066X.60.1.27|s2cid=1676488 |url=http://pdfs.semanticscholar.org/76f4/b90cf2eab3829c86b489bfdc372c1e735616.pdf |archive-url=https://web.archive.org/web/20190226012954/http://pdfs.semanticscholar.org/76f4/b90cf2eab3829c86b489bfdc372c1e735616.pdf |url-status=dead |archive-date=2019-02-26 }} |
*{{cite journal |vauthors=Helms JE, Jernigan M, Mascher J |title=The meaning of race in psychology and how to change it: a methodological perspective |journal=The American Psychologist |volume=60 |issue=1 |pages=27–36 |date=January 2005 |pmid=15641919 |doi=10.1037/0003-066X.60.1.27|s2cid=1676488 |url=http://pdfs.semanticscholar.org/76f4/b90cf2eab3829c86b489bfdc372c1e735616.pdf |archive-url=https://web.archive.org/web/20190226012954/http://pdfs.semanticscholar.org/76f4/b90cf2eab3829c86b489bfdc372c1e735616.pdf |url-status=dead |archive-date=2019-02-26 }} |
||
*{{cite journal |
*{{cite journal |vauthors=Keita SO, Kittles RA, Royal CD, etal |title=Conceptualizing human variation |journal=Nature Genetics |volume=36 |issue=11 Suppl |pages=S17–20 |date=November 2004 |pmid=15507998 |doi=10.1038/ng1455|doi-access=free }} |
||
*{{cite book |title=Revisiting Race in a Genomic Age |editor1-last=Koenig |editor1-first=Barbara A. |editor2-last=Lee |editor2-first=Sandra Soo-jin |editor3-last=Richardson |editor3-first=Sarah S. |date=2008 |publisher=Rutgers University Press |location=New Brunswick (NJ) |isbn=978-0-8135-4324-6}} This review of current research includes chapters by Jonathan Marks, John Dupré, Sally Haslanger, Deborah A. Bolnick, Marcus W. Feldman, Richard C. Lewontin, Sarah K. Tate, David B. Goldstein, Jonathan Kahn, Duana Fullwiley, Molly J. Dingel, Barbara A. Koenig, Mark D. Shriver, Rick A. Kittles, Henry T. Greely, Kimberly Tallbear, Alondra Nelson, Pamela Sankar, Sally Lehrman, Jenny Reardon, Jacqueline Stevens, and Sandra Soo-Jin Lee. |
*{{cite book |title=Revisiting Race in a Genomic Age |editor1-last=Koenig |editor1-first=Barbara A. |editor2-last=Lee |editor2-first=Sandra Soo-jin |editor3-last=Richardson |editor3-first=Sarah S. |date=2008 |publisher=Rutgers University Press |location=New Brunswick (NJ) |isbn=978-0-8135-4324-6}} This review of current research includes chapters by Jonathan Marks, John Dupré, Sally Haslanger, Deborah A. Bolnick, Marcus W. Feldman, Richard C. Lewontin, Sarah K. Tate, David B. Goldstein, Jonathan Kahn, Duana Fullwiley, Molly J. Dingel, Barbara A. Koenig, Mark D. Shriver, Rick A. Kittles, Henry T. Greely, Kimberly Tallbear, [[Alondra Nelson]], Pamela Sankar, Sally Lehrman, Jenny Reardon, Jacqueline Stevens, and Sandra Soo-Jin Lee. |
||
*{{Cite journal |last1=Lieberman |first1=Leonard |last2=Kirk |first2=Rodney C. |last3=Corcoran |first3=Michael |year=2003 |title=The Decline of Race in American Physical Anthropology |journal=Przegląd Antropologiczny – Anthropological Review |issn=0033-2003 |volume=66 |pages=3–21 |url=http://www.staff.amu.edu.pl/~anthro/pdf/paar/vol066/01lieb.pdf |access-date=2010-09-12 |archive-url=https://web.archive.org/web/20110608131024/http://www.staff.amu.edu.pl/~anthro/pdf/paar/vol066/01lieb.pdf |archive-date=2011-06-08 |url-status=dead }} |
*{{Cite journal |last1=Lieberman |first1=Leonard |last2=Kirk |first2=Rodney C. |last3=Corcoran |first3=Michael |year=2003 |title=The Decline of Race in American Physical Anthropology |journal=Przegląd Antropologiczny – Anthropological Review |issn=0033-2003 |volume=66 |pages=3–21 |url=http://www.staff.amu.edu.pl/~anthro/pdf/paar/vol066/01lieb.pdf |access-date=2010-09-12 |archive-url=https://web.archive.org/web/20110608131024/http://www.staff.amu.edu.pl/~anthro/pdf/paar/vol066/01lieb.pdf |archive-date=2011-06-08 |url-status=dead }} |
||
*{{cite journal |vauthors=Long JC, Kittles RA |title=Human genetic diversity and the nonexistence of biological races |journal=Human Biology |volume=75 |issue=4 |pages=449–71 |date=August 2003 |pmid=14655871 |doi=10.1353/hub.2003.0058|s2cid=26108602 }} |
*{{cite journal |vauthors=Long JC, Kittles RA |title=Human genetic diversity and the nonexistence of biological races |journal=Human Biology |volume=75 |issue=4 |pages=449–71 |date=August 2003 |pmid=14655871 |doi=10.1353/hub.2003.0058|s2cid=26108602 }} |
||
*{{cite journal |doi=10.1046/j.1523-1739.1996.10041115.x |title=Phylogeographic Subspecies Recognition in Leopards (Panthera pardus): Molecular Genetic Variation |year=1996 |last1=Miththapala |first1=Sriyanie |last2=Seidensticker |first2=John |last3=O'Brien |first3=Stephen J. |journal=Conservation Biology |volume=10 |pages=1115–1132 |issue=4}} |
*{{cite journal |doi=10.1046/j.1523-1739.1996.10041115.x |title=Phylogeographic Subspecies Recognition in Leopards (Panthera pardus): Molecular Genetic Variation |year=1996 |last1=Miththapala |first1=Sriyanie |last2=Seidensticker |first2=John |last3=O'Brien |first3=Stephen J. |journal=Conservation Biology |volume=10 |pages=1115–1132 |issue=4|bibcode=1996ConBi..10.1115M }} |
||
*{{cite journal |vauthors=Ossorio P, Duster T |title=Race and genetics: controversies in biomedical, behavioral, and forensic sciences |journal=The American Psychologist |volume=60 |issue=1 |pages=115–28 |date=January 2005 |pmid=15641926 |doi=10.1037/0003-066X.60.1.115}} |
*{{cite journal |vauthors=Ossorio P, Duster T |title=Race and genetics: controversies in biomedical, behavioral, and forensic sciences |journal=The American Psychologist |volume=60 |issue=1 |pages=115–28 |date=January 2005 |pmid=15641926 |doi=10.1037/0003-066X.60.1.115}} |
||
*{{cite journal |vauthors=Parra EJ, Kittles RA, Shriver MD |title=Implications of correlations between skin color and genetic ancestry for biomedical research |journal=Nature Genetics |volume=36 |issue=11 Suppl |pages=S54–60 |date=November 2004 |pmid=15508005 |doi=10.1038/ng1440|doi-access=free }} |
*{{cite journal |vauthors=Parra EJ, Kittles RA, Shriver MD |title=Implications of correlations between skin color and genetic ancestry for biomedical research |journal=Nature Genetics |volume=36 |issue=11 Suppl |pages=S54–60 |date=November 2004 |pmid=15508005 |doi=10.1038/ng1440|doi-access=free }} |
||
*{{cite journal |
*{{cite journal |vauthors=Sawyer SL, Mukherjee N, Pakstis AJ, etal |title=Linkage disequilibrium patterns vary substantially among populations |journal=European Journal of Human Genetics |volume=13 |issue=5 |pages=677–86 |date=May 2005 |pmid=15657612 |doi=10.1038/sj.ejhg.5201368|doi-access=free }} |
||
*{{cite journal |vauthors=Rohde DL, Olson S, Chang JT |title=Modelling the recent common ancestry of all living humans |journal=Nature |volume=431 |issue=7008 |pages=562–6 |date=September 2004 |pmid=15457259 |doi=10.1038/nature02842|bibcode = 2004Natur.431..562R |citeseerx=10.1.1.78.8467 |s2cid=3563900 }} |
*{{cite journal |vauthors=Rohde DL, Olson S, Chang JT |title=Modelling the recent common ancestry of all living humans |journal=Nature |volume=431 |issue=7008 |pages=562–6 |date=September 2004 |pmid=15457259 |doi=10.1038/nature02842|bibcode = 2004Natur.431..562R |citeseerx=10.1.1.78.8467 |s2cid=3563900 }} |
||
*{{cite journal |vauthors=Serre D, Pääbo S |title=Evidence for Gradients of Human Genetic Diversity Within and Among Continents |journal=Genome Research |volume=14 |issue=9 |pages=1679–85 |date=September 2004 |pmid=15342553 |pmc=515312 |doi=10.1101/gr.2529604}} |
*{{cite journal |vauthors=Serre D, Pääbo S |title=Evidence for Gradients of Human Genetic Diversity Within and Among Continents |journal=Genome Research |volume=14 |issue=9 |pages=1679–85 |date=September 2004 |pmid=15342553 |pmc=515312 |doi=10.1101/gr.2529604}} |
||
| Line 196: | Line 214: | ||
{{DEFAULTSORT:Race And Genetics}} |
{{DEFAULTSORT:Race And Genetics}} |
||
[[Category:Genetic genealogy]] |
[[Category:Genetic genealogy]] |
||
[[Category:Race (human categorization) |
[[Category:Race (human categorization)]] |
||
[[Category:Human population genetics]] |
[[Category:Human population genetics]] |
||
[[Category:Biological anthropology]] |
[[Category:Biological anthropology]] |
||
Latest revision as of 14:54, 23 August 2024
| Race |
|---|
| History |
| Society |
| Race and... |
| By location |
| Related topics |
Researchers have investigated the relationship between race and genetics as part of efforts to understand how biology may or may not contribute to human racial categorization. Today, the consensus among scientists is that race is a social construct, and that using it as a proxy for genetic differences among populations is misleading.[1][2]
Many constructions of race are associated with phenotypical traits and geographic ancestry, and scholars like Carl Linnaeus have proposed scientific models for the organization of race since at least the 18th century. Following the discovery of Mendelian genetics and the mapping of the human genome, questions about the biology of race have often been framed in terms of genetics.[3] A wide range of research methods have been employed to examine patterns of human variation and their relations to ancestry and racial groups, including studies of individual traits,[4] studies of large populations and genetic clusters,[5] and studies of genetic risk factors for disease.[6]
Research into race and genetics has also been criticized as emerging from, or contributing to, scientific racism. Genetic studies of traits and populations have been used to justify social inequalities associated with race,[7] despite the fact that patterns of human variation have been shown to be mostly clinal,[8] with human genetic code being approximately 99.6%-99.9% identical between individuals and without clear boundaries between groups.[9]
Some researchers have argued that race can act as a proxy for genetic ancestry because individuals of the same racial category may share a common ancestry, but this view has fallen increasingly out of favor among experts.[2][10] The mainstream view is that it is necessary to distinguish between biology and the social, political, cultural, and economic factors that contribute to conceptions of race.[11][12]
Phenotype may have a tangential connection to DNA, but it is still only a rough proxy that would omit various other genetic information.[2][13][14] Today, in a somewhat similar way that "gender" is differentiated from the more clear "biological sex", scientists state that potentially "race" / phenotype can be differentiated from the more clear "ancestry".[15] However, this system has also still come under scrutiny as it may fall into the same problems – which would be large, vague groupings with little genetic value.[16]
Übersicht
[edit]The concept of race
[edit]The concept of "race" as a classification system of humans based on visible physical characteristics emerged over the last five centuries, influenced by European colonialism.[11][17] However, there is widespread evidence of what would be described in modern terms as racial consciousness throughout the entirety of recorded history. For example, in Ancient Egypt there were four broad racial divisions of human beings: Egyptians, Asiatics, Libyans, and Nubians.[18] There was also Aristotle of Ancient Greece, who once wrote: "The peoples of Asia... lack spirit, so that they are in continuous subjection and slavery."[19] The concept has manifested in different forms based on social conditions of a particular group, often used to justify unequal treatment. Early influential attempts to classify humans into discrete races include 4 races in Carl Linnaeus's Systema Naturae (Homo europaeus, asiaticus, americanus, and afer)[20][21] and 5 races in Johann Friedrich Blumenbach's On the Natural Variety of Mankind.[22] Notably, over the next centuries, scholars argued for anywhere from 3 to more than 60 race categories.[23] Race concepts have changed within a society over time; for example, in the United States social and legal designations of "White" have been inconsistently applied to Native Americans, Arab Americans, and Asian Americans, among other groups (See main article: Definitions of whiteness in the United States). Race categories also vary worldwide; for example, the same person might be perceived as belonging to a different category in the United States versus Brazil.[24] Because of the arbitrariness inherent in the concept of race, it is difficult to relate it to biology in a straightforward way.
Race and human genetic variation
[edit]There is broad consensus across the biological and social sciences that race is a social construct, not an accurate representation of human genetic variation.[25][9] As more progress has been made on sequencing the human genome, it has been found that any two humans will share an average of 99.35% of their DNA based on the approximately 3.1 billion haploid base pairs.[26][27] However, this number should be understood as an average, any two specific individuals can have their genomes differ by more or less than 0.65%. Additionally, this average is an estimate, subject to change as additional sequences are discovered and populations sampled. In 2010, the genome of Craig Venter was found to differ by an estimated 1.59% from a reference genome created by the National Center for Biotechnology Information.[28]
We nonetheless see wide individual variation in phenotype, which arises from both genetic differences and complex gene-environment interactions. The vast majority of this genetic variation occurs within groups; very little genetic variation differentiates between groups.[5] Crucially, the between-group genetic differences that do exist do not map onto socially recognized categories of race. Furthermore, although human populations show some genetic clustering across geographic space, human genetic variation is "clinal", or continuous.[11][9] This, in addition to the fact that different traits vary on different clines, makes it impossible to draw discrete genetic boundaries around human groups. Finally, insights from ancient DNA are revealing that no human population is "pure" – all populations represent a long history of migration and mixing.[29]
Sources of human genetic variation
[edit]Genetic variation arises from mutations, from natural selection, migration between populations (gene flow) and from the reshuffling of genes through sexual reproduction.[30] Mutations lead to a change in the DNA structure, as the order of the bases are rearranged. Resultantly, different polypeptide proteins are coded. Some mutations may be positive and can help the individual survive more effectively in their environment. Mutation is counteracted by natural selection and by genetic drift; note too the founder effect, when a small number of initial founders establish a population which hence starts with a correspondingly small degree of genetic variation.[31] Epigenetic inheritance involves heritable changes in phenotype (appearance) or gene expression caused by mechanisms other than changes in the DNA sequence.[32]
Human phenotypes are highly polygenic (dependent on interaction by many genes) and are influenced by environment as well as by genetics.
Nucleotide diversity is based on single mutations, single nucleotide polymorphisms (SNPs). The nucleotide diversity between humans is about 0.1 percent (one difference per one thousand nucleotides between two humans chosen at random). This amounts to approximately three million SNPs (since the human genome has about three billion nucleotides). There are an estimated ten million SNPs in the human population.[33]
Research has shown that non-SNP (structural) variation accounts for more human genetic variation than single nucleotide diversity. Structural variation includes copy-number variation and results from deletions, inversions, insertions and duplications. It is estimated that approximately 0.4 to 0.6 percent of the genomes of unrelated people differ.[9][34]
Genetic basis for race
[edit]Much scientific research has been organized around the question of whether or not there is genetic basis for race. In Luigi Luca Cavalli-Sforza's book (circa 1994) "The History and Geography of Human Genes"[35] he writes, "From a scientific point of view, the concept of race has failed to obtain any consensus; none is likely, given the gradual variation in existence. It may be objected that the racial stereotypes have a consistency that allows even the layman to classify individuals. However, the major stereotypes, all based on skin color, hair color and form, and facial traits, reflect superficial differences that are not confirmed by deeper analysis with more reliable genetic traits and whose origin dates from recent evolution mostly under the effect of climate and perhaps sexual selection".
In 2018 geneticist David Reich reaffirmed the conclusion that the traditional views which assert a biological basis for race are wrong:
Today, many people assume that humans can be grouped biologically into "primeval" groups, corresponding to our notion of "races"... But this long-held view about "race" has just in the last years been proven wrong.
— David Reich, Who We Are and How We Got Here, (Introduction, pg. xxiv).
In 1956, some scientists proposed that race may be similar to dog breeds within dogs. However, this theory has since been discarded, with one of the main reasons being that purebred dogs have been specifically bred artificially, whereas human races developed organically.[36] Furthermore, the genetic variation between purebred dog breeds is far greater than that of human populations. Dog-breed intervariation is roughly 27.5%, whereas human populations inter-variation is only at 10-15.6%.[37][38][39][40] Including non purebreds would substantially decrease the 27.5% genetic variance, however. Mammal taxonomy is rarely defined by genetic variance alone.
Research methods
[edit]Scientists investigating human variation have used a series of methods to characterize how different populations vary.
Early studies of traits, proteins, and genes
[edit]Early racial classification attempts measured surface traits, particularly skin color, hair color and texture, eye color, and head size and shape. (Measurements of the latter through craniometry were repeatedly discredited in the late 19th and mid-20th centuries due to a lack of correlation of phenotypic traits with racial categorization.[41]) In actuality, biological adaptation plays the biggest role in these bodily features and skin type. A relative handful of genes accounts for the inherited factors shaping a person's appearance.[42][43] Humans have an estimated 19,000–20,000 human protein-coding genes.[44] Richard Sturm and David Duffy describe 11 genes that affect skin pigmentation and explain most variations in human skin color, the most significant of which are MC1R, ASIP, OCA2, and TYR.[45] There is evidence that as many as 16 different genes could be responsible for eye color in humans; however, the main two genes associated with eye color variation are OCA2 and HERC2, and both are localized in chromosome 15.[46]
Analysis of blood proteins and between-group genetics
[edit]

Before the discovery of DNA, scientists used blood proteins (the human blood group systems) to study human genetic variation. Research by Ludwik and Hanka Herschfeld during World War I found that the incidence of blood groups A and B differed by region; for example, among Europeans 15 percent were group B and 40 percent group A. Eastern Europeans and Russians had a higher incidence of group B; people from India had the greatest incidence. The Herschfelds concluded that humans comprised two "biochemical races", originating separately. It was hypothesized that these two races later mixed, resulting in the patterns of groups A and B. This was one of the first theories of racial differences to include the idea that human variation did not correlate with genetic variation. It was expected that groups with similar proportions of blood groups would be more closely related, but instead it was often found that groups separated by great distances (such as those from Madagascar and Russia), had similar incidences.[47] It was later discovered that the ABO blood group system is not just common to humans, but shared with other primates,[48] and likely predates all human groups.[49]
In 1972, Richard Lewontin performed a FST statistical analysis using 17 markers (including blood-group proteins). He found that the majority of genetic differences between humans (85.4 percent) were found within a population, 8.3 percent were found between populations within a race and 6.3 percent were found to differentiate races (Caucasian, African, Mongoloid, South Asian Aborigines, Amerinds, Oceanians, and Australian Aborigines in his study). Since then, other analyses have found FST values of 6–10 percent between continental human groups, 5–15 percent between different populations on the same continent and 75–85 percent within populations.[50][51][52][53][54] This view has been affirmed by the American Anthropological Association and the American Association of Physical Anthropologists since.[55]
Critiques of blood protein analysis
[edit]While acknowledging Lewontin's observation that humans are genetically homogeneous, A. W. F. Edwards in his 2003 paper "Human Genetic Diversity: Lewontin's Fallacy" argued that information distinguishing populations from each other is hidden in the correlation structure of allele frequencies, making it possible to classify individuals using mathematical techniques. Edwards argued that even if the probability of misclassifying an individual based on a single genetic marker is as high as 30 percent (as Lewontin reported in 1972), the misclassification probability nears zero if enough genetic markers are studied simultaneously. Edwards saw Lewontin's argument as based on a political stance, denying biological differences to argue for social equality.[56] Edwards' paper is reprinted, commented upon by experts such as Noah Rosenberg, and given further context in an interview with philosopher of science Rasmus Grønfeldt Winther in a recent anthology.[57]
As referred to before, Edwards criticises Lewontin's paper as he took 17 different traits and analysed them independently, without looking at them in conjunction with any other protein. Thus, it would have been fairly convenient for Lewontin to come up with the conclusion that racial naturalism is not tenable, according to his argument.[58] Sesardic also strengthened Edwards' view, as he used an illustration referring to squares and triangles, and showed that if you look at one trait in isolation, then it will most likely be a bad predicator of which group the individual belongs to.[59] In contrast, in a 2014 paper, reprinted in the 2018 Edwards Cambridge University Press volume, Rasmus Grønfeldt Winther argues that "Lewontin's Fallacy" is effectively a misnomer, as there really are two different sets of methods and questions at play in studying the genomic population structure of our species: "variance partitioning" and "clustering analysis." According to Winther, they are "two sides of the same mathematics coin" and neither "necessarily implies anything about the reality of human groups."[60]
Current studies of population genetics
[edit]This section needs to be updated. The reason given is: All references are more than 13 years old. (December 2022) |
Researchers currently use genetic testing, which may involve hundreds (or thousands) of genetic markers or the entire genome.
Structure
[edit]
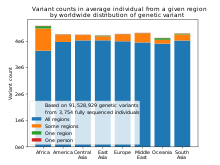
Several methods to examine and quantify genetic subgroups exist, including cluster and principal components analysis. Genetic markers from individuals are examined to find a population's genetic structure. While subgroups overlap when examining variants of one marker only, when a number of markers are examined different subgroups have different average genetic structure. An individual may be described as belonging to several subgroups. These subgroups may be more or less distinct, depending on how much overlap there is with other subgroups.[62]
In cluster analysis, the number of clusters to search for K is determined in advance; how distinct the clusters are varies.
The results obtained from cluster analyses depend on several factors:
- A large number of genetic markers studied facilitates finding distinct clusters.[63]
- Some genetic markers vary more than others, so fewer are required to find distinct clusters.[5] Ancestry-informative markers exhibit substantially different frequencies between populations from different geographical regions. Using AIMs, scientists can determine a person's ancestral continent of origin based solely on their DNA. AIMs can also be used to determine someone's admixture proportions.[64]
- The more individuals studied, the easier it becomes to detect distinct clusters (statistical noise is reduced).[5]
- Low genetic variation makes it more difficult to find distinct clusters.[5] Greater geographic distance generally increases genetic variation, making identifying clusters easier.[65]
- A similar cluster structure is seen with different genetic markers when the number of genetic markers included is sufficiently large. The clustering structure obtained with different statistical techniques is similar. A similar cluster structure is found in the original sample with a subsample of the original sample.[66]
Recent studies have been published using an increasing number of genetic markers.[5][66][67][68][69][70]
Focus on study of structure has been criticized for giving the general public a misleading impression of human genetic variation, obscuring the general finding that genetic variants which are limited to one region tend to be rare within that region, variants that are common within a region tend to be shared across the globe, and most differences between individuals, whether they come from the same region or different regions, are due to global variants.[71]
Distance
[edit]Genetic distance is genetic divergence between species or populations of a species. It may compare the genetic similarity of related species, such as humans and chimpanzees. Within a species, genetic distance measures divergence between subgroups. Genetic distance significantly correlates to geographic distance between populations, a phenomenon sometimes known as "isolation by distance".[72] Genetic distance may be the result of physical boundaries restricting gene flow such as islands, deserts, mountains or forests. Genetic distance is measured by the fixation index (FST). FST is the correlation of randomly chosen alleles in a subgroup to a larger population. It is often expressed as a proportion of genetic diversity. This comparison of genetic variability within (and between) populations is used in population genetics. The values range from 0 to 1; zero indicates the two populations are freely interbreeding, and one would indicate that two populations are separate.
Many studies place the average FST distance between human races at about 0.125. Henry Harpending argued that this value implies on a world scale a "kinship between two individuals of the same human population is equivalent to kinship between grandparent and grandchild or between half siblings". In fact, the formulas derived in Harpending's paper in the "Kinship in a subdivided population" section imply that two unrelated individuals of the same race have a higher coefficient of kinship (0.125) than an individual and their mixed race half-sibling (0.109).[73]
Critiques of FST
[edit]While acknowledging that FST remains useful, a number of scientists have written about other approaches to characterizing human genetic variation.[74][75][76] Long & Kittles (2009) stated that FST failed to identify important variation and that when the analysis includes only humans, FST = 0.119, but adding chimpanzees increases it only to FST = 0.183.[74] Mountain & Risch (2004) argued that an FST estimate of 0.10–0.15 does not rule out a genetic basis for phenotypic differences between groups and that a low FST estimate implies little about the degree to which genes contribute to between-group differences.[75] Pearse & Crandall 2004 wrote that FST figures cannot distinguish between a situation of high migration between populations with a long divergence time, and one of a relatively recent shared history but no ongoing gene flow.[76] In their 2015 article, Keith Hunley, Graciela Cabana, and Jeffrey Long (who had previously criticized Lewontin's statistical methodology with Rick Kittles[55]) recalculate the apportionment of human diversity using a more complex model than Lewontin and his successors. They conclude: "In sum, we concur with Lewontin's conclusion that Western-based racial classifications have no taxonomic significance, and we hope that this research, which takes into account our current understanding of the structure of human diversity, places his seminal finding on firmer evolutionary footing."[77]
Anthropologists (such as C. Loring Brace),[78] philosopher Jonathan Kaplan and geneticist Joseph Graves[79] have argued that while it is possible to find biological and genetic variation roughly corresponding to race, this is true for almost all geographically distinct populations: the cluster structure of genetic data is dependent on the initial hypotheses of the researcher and the populations sampled. When one samples continental groups, the clusters become continental; with other sampling patterns, the clusters would be different. Weiss and Fullerton note that if one sampled only Icelanders, Mayans and Maoris, three distinct clusters would form; all other populations would be composed of genetic admixtures of Maori, Icelandic and Mayan material.[80] Kaplan therefore concludes that, while differences in particular allele frequencies can be used to identify populations that loosely correspond to the racial categories common in Western social discourse, the differences are of no more biological significance than the differences found between any human populations (e.g., the Spanish and Portuguese).[81]
Historical and geographical analyses
[edit]Current-population genetic structure does not imply that differing clusters or components indicate only one ancestral home per group; for example, a genetic cluster in the US comprises Hispanics with European, Native American and African ancestry.[63]
Geographic analyses attempt to identify places of origin, their relative importance and possible causes of genetic variation in an area. The results can be presented as maps showing genetic variation. Cavalli-Sforza and colleagues argue that if genetic variations are investigated, they often correspond to population migrations due to new sources of food, improved transportation or shifts in political power. For example, in Europe the most significant direction of genetic variation corresponds to the spread of agriculture from the Middle East to Europe between 10,000 and 6,000 years ago.[82] Such geographic analysis works best in the absence of recent large-scale, rapid migrations.
Historic analyses use differences in genetic variation (measured by genetic distance) as a molecular clock indicating the evolutionary relation of species or groups, and can be used to create evolutionary trees reconstructing population separations.[82]
Results of genetic-ancestry research are supported if they agree with research results from other fields, such as linguistics or archeology.[82] Cavalli-Sforza and colleagues have argued that there is a correspondence between language families found in linguistic research and the population tree they found in their 1994 study. There are generally shorter genetic distances between populations using languages from the same language family. Exceptions to this rule are also found, for example Sami, who are genetically associated with populations speaking languages from other language families. The Sami speak a Uralic language, but are genetically primarily European. This is argued to have resulted from migration (and interbreeding) with Europeans while retaining their original language. Agreement also exists between research dates in archeology and those calculated using genetic distance.[5][82]
Self-identification studies
[edit]Jorde and Wooding found that while clusters from genetic markers were correlated with some traditional concepts of race, the correlations were imperfect and imprecise due to the continuous and overlapping nature of genetic variation, noting that ancestry, which can be accurately determined, is not equivalent to the concept of race.[33]
A 2005 study by Tang and colleagues used 326 genetic markers to determine genetic clusters. The 3,636 subjects, from the United States and Taiwan, self-identified as belonging to white, African American, East Asian or Hispanic ethnic groups. The study found "nearly perfect correspondence between genetic cluster and SIRE for major ethnic groups living in the United States, with a discrepancy rate of only 0.14 percent".[63] Paschou et al. found "essentially perfect" agreement between 51 self-identified populations of origin and the population's genetic structure, using 650,000 genetic markers. Selecting for informative genetic markers allowed a reduction to less than 650, while retaining near-total accuracy.[83]
Correspondence between genetic clusters in a population (such as the current US population) and self-identified race or ethnic groups does not mean that such a cluster (or group) corresponds to only one ethnic group. African Americans have an estimated 20–25-percent European genetic admixture; Hispanics have European, Native American and African ancestry.[63] In Brazil there has been extensive admixture between Europeans, Amerindians and Africans. As a result, skin color differences within the population are not gradual, and there are relatively weak associations between self-reported race and African ancestry.[84][85] Ethnoracial self- classification in Brazilians is certainly not random with respect to genome individual ancestry, but the strength of the association between the phenotype and median proportion of African ancestry varies largely across population.[86]
Critique of genetic-distance studies and clusters
[edit]
Genetic distances generally increase continually with geographic distance, which makes a dividing line arbitrary. Any two neighboring settlements will exhibit some genetic difference from each other, which could be defined as a race. Therefore, attempts to classify races impose an artificial discontinuity on a naturally occurring phenomenon. This explains why studies on population genetic structure yield varying results, depending on methodology.[87]
Rosenberg and colleagues (2005) have argued, based on cluster analysis of the 52 populations in the Human Genetic Diversity Panel, that populations do not always vary continuously and a population's genetic structure is consistent if enough genetic markers (and subjects) are included.
Examination of the relationship between genetic and geographic distance supports a view in which the clusters arise not as an artifact of the sampling scheme, but from small discontinuous jumps in genetic distance for most population pairs on opposite sides of geographic barriers, in comparison with genetic distance for pairs on the same side. Thus, analysis of the 993-locus dataset corroborates our earlier results: if enough markers are used with a sufficiently large worldwide sample, individuals can be partitioned into genetic clusters that match major geographic subdivisions of the globe, with some individuals from intermediate geographic locations having mixed membership in the clusters that correspond to neighboring regions.
They also wrote, regarding a model with five clusters corresponding to Africa, Eurasia (Europe, Middle East, and Central/South Asia), East Asia, Oceania, and the Americas:
For population pairs from the same cluster, as geographic distance increases, genetic distance increases in a linear manner, consistent with a clinal population structure. However, for pairs from different clusters, genetic distance is generally larger than that between intracluster pairs that have the same geographic distance. For example, genetic distances for population pairs with one population in Eurasia and the other in East Asia are greater than those for pairs at equivalent geographic distance within Eurasia or within East Asia. Loosely speaking, it is these small discontinuous jumps in genetic distance—across oceans, the Himalayas, and the Sahara—that provide the basis for the ability of STRUCTURE to identify clusters that correspond to geographic regions.[66]
This applies to populations in their ancestral homes when migrations and gene flow were slow; large, rapid migrations exhibit different characteristics. Tang and colleagues (2004) wrote, "we detected only modest genetic differentiation between different current geographic locales within each race/ethnicity group. Thus, ancient geographic ancestry, which is highly correlated with self-identified race/ethnicity—as opposed to current residence—is the major determinant of genetic structure in the U.S. population".[63]

Cluster analysis has been criticized because the number of clusters to search for is decided in advance, with different values possible (although with varying degrees of probability).[88] Principal component analysis does not decide in advance how many components for which to search.[89]
The 2002 study by Rosenberg et al. exemplifies why meanings of these clusterings can be disputable, though the study shows that at the K=5 cluster analysis, genetic clusterings roughly map onto each of the five major geographical regions.[5] Similar results were gathered in further studies in 2005.[90]
Critique of ancestry-informative markers
[edit]Ancestry-informative markers (AIMs) are a genealogy tracing technology that has come under much criticism due to its reliance on reference populations. In a 2015 article, Troy Duster outlines how contemporary technology allows the tracing of ancestral lineage but along only the lines of one maternal and one paternal line. That is, of 64 total great-great-great-great-grandparents, only one from each parent is identified, implying the other 62 ancestors are ignored in tracing efforts.[91] Furthermore, the 'reference populations' used as markers for membership of a particular group are designated arbitrarily and contemporarily. In other words, using populations who currently reside in given places as references for certain races and ethnic groups is unreliable due to the demographic changes which have occurred over many centuries in those places. Furthermore, ancestry-informative markers being widely shared among the whole human population, it is their frequency which is tested, not their mere absence/presence. A threshold of relative frequency has, therefore, to be set. According to Duster, the criteria for setting such thresholds are a trade secret of the companies marketing the tests. Thus, we cannot say anything conclusive on whether they are appropriate. Results of AIMs are extremely sensitive to where this bar is set.[92] Given that many genetic traits are found to be very similar amid many different populations, the designated threshold frequencies are very important. This can also lead to mistakes, given that many populations may share the same patterns, if not exactly the same genes. "This means that someone from Bulgaria whose ancestors go back to the fifteenth century could (and sometime does) map as partly 'Native American'".[91] This happens because AIMs rely on a '100% purity' assumption of reference populations. That is, they assume that a pattern of traits would ideally be a necessary and sufficient condition for assigning an individual to an ancestral reference populations.
Race, genetics, and medicine
[edit]There are certain statistical differences between racial groups in susceptibility to certain diseases.[93] Genes change in response to local diseases; for example, people who are Duffy-negative tend to have a higher resistance to malaria. The Duffy negative phenotype is highly frequent in central Africa and the frequency decreases with distance away from Central Africa, with higher frequencies in global populations with high degrees of recent African immigration. This suggests that the Duffy negative genotype evolved in Sub-Saharan Africa and was subsequently positively selected for in the Malaria endemic zone.[94] A number of genetic conditions prevalent in malaria-endemic areas may provide genetic resistance to malaria, including sickle cell disease, thalassaemias and glucose-6-phosphate dehydrogenase. Cystic fibrosis is the most common life-limiting autosomal recessive disease among people of European ancestry; a hypothesized heterozygote advantage, providing resistance to diseases earlier common in Europe, has been challenged.[95] Scientists Michael Yudell, Dorothy Roberts, Rob DeSalle, and Sarah Tishkoff argue that using these associations in the practice of medicine has led doctors to overlook or misidentify disease: "For example, hemoglobinopathies can be misdiagnosed because of the identification of sickle-cell as a 'Black' disease and thalassemia as a 'Mediterranean' disease. Cystic fibrosis is underdiagnosed in populations of African ancestry, because it is thought of as a 'White' disease."[25]
Information about a person's population of origin may aid in diagnosis, and adverse drug responses may vary by group.[5][dubious – discuss] Because of the correlation between self-identified race and genetic clusters, medical treatments influenced by genetics have varying rates of success between self-defined racial groups.[96] For this reason, some physicians[who?] consider a patient's race in choosing the most effective treatment,[97] and some drugs are marketed with race-specific instructions.[98] Jorde and Wooding (2004) have argued that because of genetic variation within racial groups, when "it finally becomes feasible and available, individual genetic assessment of relevant genes will probably prove more useful than race in medical decision making". However, race continues to be a factor when examining groups (such as epidemiologic research).[33] Some doctors and scientists such as geneticist Neil Risch argue that using self-identified race as a proxy for ancestry is necessary to be able to get a sufficiently broad sample of different ancestral populations, and in turn to be able to provide health care that is tailored to the needs of minority groups.[99]
Usage in scientific journals
[edit]Some scientific journals have addressed previous methodological errors by requiring more rigorous scrutiny of population variables. Since 2000, Nature Genetics requires its authors to "explain why they make use of particular ethnic groups or populations, and how classification was achieved". Editors of Nature Genetics say that "[they] hope that this will raise awareness and inspire more rigorous designs of genetic and epidemiological studies".[100]
A 2021 study that examined over 11,000 papers from 1949 to 2018 in The American Journal of Human Genetics, found that "race" was used in only 5% of papers published in the last decade, down from 22% in the first. Together with an increase in use of the terms "ethnicity," "ancestry," and location-based terms, it suggests that human geneticists have mostly abandoned the term "race."[101]
Gene-environment interactions
[edit]Lorusso and Bacchini[6] argue that self-identified race is of greater use in medicine as it correlates strongly with risk-related exposomes that are potentially heritable when they become embodied in the epigenome. They summarise evidence of the link between racial discrimination and health outcomes due to poorer food quality, access to healthcare, housing conditions, education, access to information, exposure to infectious agents and toxic substances, and material scarcity. They also cite evidence that this process can work positively – for example, the psychological advantage of perceiving oneself at the top of a social hierarchy is linked to improved health. However they caution that the effects of discrimination do not offer a complete explanation for differential rates of disease and risk factors between racial groups, and the employment of self-identified race has the potential to reinforce racial inequalities.
Objections to racial naturalism
[edit]Racial naturalism is the view that racial classifications are grounded in objective patterns of genetic similarities and differences. Proponents of this view have justified it using the scientific evidence described above. However, this view is controversial and philosophers[102] of race have put forward four main objections to it.
Semantic objections, such as the discreteness objection, argue that the human populations picked out in population-genetic research are not races and do not correspond to what "race" means in the United States. "The discreteness objection does not require there to be no genetic admixture in the human species in order for there to be US 'racial groups' ... rather ... what the objection claims is that membership in US racial groups is different from membership in continental populations. ... Thus, strictly speaking, Blacks are not identical to Africans, Whites are not identical to Eurasians, Asians are not identical to East Asians and so forth."[103] Therefore, it could be argued that scientific research is not really about race.
The next two objections, are metaphysical objections which argue that even if the semantic objections fail, human genetic clustering results do not support the biological reality of race. The 'very important objection' stipulates that races in the US definition fail to be important to biology, in the sense that continental populations do not form biological subspecies. The 'objectively real objection' states that "US racial groups are not biologically real because they are not objectively real in the sense of existing independently of human interest, belief, or some other mental state of humans."[104] Racial naturalists, such as Quayshawn Spencer, have responded to each of these objections with counter-arguments. There are also methodological critics who reject racial naturalism because of concerns relating to the experimental design, execution, or interpretation of the relevant population-genetic research.[105]
Another semantic objection is the visibility objection which refutes the claim that there are US racial groups in human population structures. Philosophers such as Joshua Glasgow and Naomi Zack believe that US racial groups cannot be defined by visible traits, such as skin colour and physical attributes: "The ancestral genetic tracking material has no effect on phenotypes, or biological traits of organisms, which would include the traits deemed racial, because the ancestral tracking genetic material plays no role in the production of proteins it is not the kind of material that 'codes' for protein production."[106][page needed] Spencer contends that certain racial discourses require visible groups, but disagrees that this is a requirement in all US racial discourse.[citation needed][undue weight? – discuss]
A different objection states that US racial groups are not biologically real because they are not objectively real in the sense of existing independently of some mental state of humans. Proponents of this second metaphysical objection include Naomi Zack and Ron Sundstrom.[106][107] Spencer argues that an entity can be both biologically real and socially constructed. Spencer states that in order to accurately capture real biological entities, social factors must also be considered.[citation needed][undue weight? – discuss]
It has been argued that knowledge of a person's race is limited in value, since people of the same race vary from one another.[33] David J. Witherspoon and colleagues have argued that when individuals are assigned to population groups, two randomly chosen individuals from different populations can resemble each other more than a randomly chosen member of their own group. They found that many thousands of genetic markers had to be used for the answer to "How often is a pair of individuals from one population genetically more dissimilar than two individuals chosen from two different populations?" to be "Never". This assumed three population groups, separated by large geographic distances (European, African and East Asian). The global human population is more complex, and studying a large number of groups would require an increased number of markers for the same answer. They conclude that "caution should be used when using geographic or genetic ancestry to make inferences about individual phenotypes",[108] and "The fact that, given enough genetic data, individuals can be correctly assigned to their populations of origin is compatible with the observation that most human genetic variation is found within populations, not between them. It is also compatible with our finding that, even when the most distinct populations are considered and hundreds of loci are used, individuals are frequently more similar to members of other populations than to members of their own population".[109]
This is similar to the conclusion reached by anthropologist Norman Sauer in a 1992 article on the ability of forensic anthropologists to assign "race" to a skeleton, based on craniofacial features and limb morphology. Sauer said, "the successful assignment of race to a skeletal specimen is not a vindication of the race concept, but rather a prediction that an individual, while alive was assigned to a particular socially constructed 'racial' category. A specimen may display features that point to African ancestry. In this country that person is likely to have been labeled Black regardless of whether or not such a race actually exists in nature".[110]
Criticism of race-based medicines
[edit]Troy Duster points out that genetics is often not the predominant determinant of disease susceptibilities, even though they might correlate with specific socially defined categories. This is because this research oftentimes lacks control for a multiplicity of socio-economic factors. He cites data collected by King and Rewers that indicates how dietary differences play a significant role in explaining variations of diabetes prevalence between populations.
Duster elaborates by putting forward the example of the Pima of Arizona, a population suffering from disproportionately high rates of diabetes. The reason for such, he argues, was not necessarily a result of the prevalence of the FABP2 gene, which is associated with insulin resistance. Rather he argues that scientists often discount the lifestyle implications under specific socio-historical contexts. For instance, near the end of the 19th century, the Pima economy was predominantly agriculture-based. However, as the European American population settles into traditionally Pima territory, the Pima lifestyles became heavily Westernised. Within three decades, the incidence of diabetes increased multiple folds. Governmental provision of free relatively high-fat food to alleviate the prevalence of poverty in the population is noted as an explanation of this phenomenon.[111]
Lorusso and Bacchini argue against the assumption that "self-identified race is a good proxy for a specific genetic ancestry"[6] on the basis that self-identified race is complex: it depends on a range of psychological, cultural and social factors, and is therefore "not a robust proxy for genetic ancestry".[112] Furthermore, they explain that an individual's self-identified race is made up of further, collectively arbitrary factors: personal opinions about what race is and the extent to which it should be taken into consideration in everyday life. Furthermore, individuals who share a genetic ancestry may differ in their racial self-identification across historical or socioeconomic contexts. From this, Lorusso and Bacchini conclude that the accuracy in the prediction of genetic ancestry on the basis of self-identification is low, specifically in racially admixed populations born out of complex ancestral histories.
See also
[edit]- List of Y-chromosome haplogroups in populations of the world
- History of anthropometry – Historical uses of anthropometry, section; 4.2 Race, identity and cranio-facial description
- Human subspecies – Classification of the human species
- Human Genetic Diversity: Lewontin's Fallacy – 2003 paper by A. W. F. Edwards
- Zionism, race and genetics
References
[edit]- ^ Using Population Descriptors in Genetics and Genomics Research: A New Framework for an Evolving Field (Consensus Study Report). National Academies of Sciences, Engineering, and Medicine. 2023. doi:10.17226/26902. ISBN 978-0-309-70065-8. PMID 36989389.
In humans, race is a socially constructed designation, a misleading and harmful surrogate for population genetic differences, and has a long history of being incorrectly identified as the major genetic reason for phenotypic differences between groups.
- ^ a b c "Researchers Need to Rethink and Justify How and Why Race, Ethnicity, and Ancestry Labels Are Used in Genetics and Genomics Research, Says New Report". National Academies of Sciences, Engineering, and Medicine. 14 March 2023.
Researchers and scientists who utilize genetic and genomic data should rethink and justify how and why they use race, ethnicity, and ancestry labels in their work, says a new National Academies of Sciences, Engineering, and Medicine report. The report says researchers should not use race as a proxy for describing human genetic variation. Race is a social concept, but it is often used in genomics and genetics research as a surrogate for describing human genetic differences, which is misleading, inaccurate, and harmful.
- ^ Goodman AH (2020). Race : are we so different?. Yolanda T. Moses, Joseph L. Jones (Second ed.). Hoboken, NJ: Wiley Blackwell. ISBN 978-1-119-47247-6. OCLC 1121420797. Archived from the original on 25 May 2021. Retrieved 8 April 2021.
- ^ Jablonski NG (2006). Skin : a natural history. Berkeley: University of California Press. ISBN 0-520-24281-5. OCLC 64592114. Archived from the original on 25 May 2021. Retrieved 8 April 2021.
- ^ a b c d e f g h i Rosenberg NA, Pritchard JK, Weber JL, Cann HM, et al. (20 December 2002). "Genetic structure of human populations". Science. 298 (5602): 2381–2385. Bibcode:2002Sci...298.2381R. doi:10.1126/science.1078311. ISSN 1095-9203. PMID 12493913. S2CID 8127224. Archived from the original on 30 April 2021. Retrieved 8 April 2021.
- ^ a b c Lorusso L, Bacchini F (August 2015). "A reconsideration of the role of self-identified races in epidemiology and biomedical research". Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 56–64. doi:10.1016/j.shpsc.2015.02.004. PMID 25791919. Archived from the original on 8 March 2021. Retrieved 8 April 2021.
- ^ Saini A (2019). Superior : the return of race science. Boston: Beacon Press. ISBN 978-0-8070-7691-0. OCLC 1091260230. Archived from the original on 8 August 2021. Retrieved 8 April 2021.
- ^ Marks J (2017). Is science racist?. Malden, MA: Polity. ISBN 978-0-7456-8921-0. OCLC 961801723. Archived from the original on 2 May 2021. Retrieved 8 April 2021.
- ^ a b c d Tishkoff SA, Kidd KK (26 October 2004). "Implications of biogeography of human populations for 'race' and medicine". Nature Genetics. Supplemental. 36 (11). Nature Portfolio: S21-7. doi:10.1038/ng1438. PMID 15507999. S2CID 1500915. Retrieved 26 June 2024.
- ^ Kaiser J (11 March 2023). "Geneticists should rethink how they use race and ethnicity, panel urges". Science.
- ^ a b c Ackermann R, Athreya S, Bolnick D, Fuentes A, et al. (2019). "AABA (AAPA) Statement on Race & Racism" (Press release). Retrieved 26 June 2024.
- ^ Bamshad M, Wooding S, Salisbury BA, Stephens JC (August 2004). "Deconstructing the relationship between genetics and race". Nature Reviews Genetics. 5 (8): 598–609. doi:10.1038/nrg1401. ISSN 1471-0056. PMID 15266342. S2CID 12378279. Archived from the original on 10 June 2021. Retrieved 8 April 2021.
- ^ Gannon M. "Race Is a Social Construct, Scientists Argue". Scientific American. Retrieved 12 March 2024.
- ^ Hesman Saey T (14 March 2023). "Why experts recommend ditching racial labels in genetic studies". ScienceNews. Retrieved 12 March 2024.
- ^ Chou V (18 April 2017). "How Science and Genetics are Reshaping the Race Debate of the 21st Century". Science in the News. Retrieved 12 March 2024.
- ^ Lewis AC (2 May 2022). "Substituting genetic ancestry for race in research? Not so fast". STAT. Retrieved 12 March 2024.
- ^ Kennedy RF, Roy CS, Goldman ML (2013). Race and Ethnicity in the Classical World. Indianapolis, Indiana: Hackett Publishing Company, Inc. p. xiii. ISBN 978-1603849944.
- ^ "Race in Ancient Egypt". www.ucl.ac.uk. Archived from the original on 31 July 2021. Retrieved 31 July 2021.
- ^ "Aristotle, Politics, Book 7, section 1327b". www.perseus.tufts.edu. Archived from the original on 31 July 2021. Retrieved 31 July 2021.
- ^ Slotkin JS (1965). Readings in Early Anthropology. London: Routledge. ISBN 9780203715215.
- ^ Linnaeus C (1758). Systema naturae. Stockholm: Laurentii Salvii. p. 532.
- ^ Blumenbach J, Bendyshe T (1795). On the natural variety of mankind.
- ^ Darwin C (1871). The Descent of Man, and Selection in Relation to Sex.
- ^ Daniel G (2006). Race and multiraciality in Brazil and the United States: converging paths?. Penn State Press.
- ^ a b Yudell M, Roberts D, DeSalle R, Tishkoff S (2016). "Taking race out of human genetics". Science. 351 (6273). American Association for the Advancement of Science: 564–565. Bibcode:2016Sci...351..564Y. doi:10.1126/science.aac4951. ISSN 0036-8075. OCLC 6005630581. PMID 26912690. S2CID 206639306. Archived from the original on 13 September 2021. Retrieved 27 June 2024.
- ^ Auton A, Abecasis GR, Altshuler DM, Durbin RM, et al. (October 2015). "A global reference for human genetic variation". Nature. 526 (7571). Nature Portfolio: 68–74. Bibcode:2015Natur.526...68T. doi:10.1038/nature15393. ISSN 1476-4687. OCLC 8521848829. PMC 4750478. PMID 26432245. Auton2015.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ Piovesan A, Chiara Pelleri M, Antonaros F, Strippoli P, et al. (February 2019). "On the length, weight and GC content of the human genome". BMC Research Notes. 12 (1). BioMed Central: 106. doi:10.1186/s13104-019-4137-z. ISSN 1756-0500. OCLC 8016439744. PMC 6391780. PMID 30813969.
- ^ Pang AW, MacDonald JR, Pinto D, Wei J, et al. (May 2010). "Towards a comprehensive structural variation map of an individual human genome". Genome Biology. 11 (5). BioMed Central: R52. doi:10.1186/gb-2010-11-5-r52. ISSN 1474-760X. OCLC 5660396679. PMC 2898065. PMID 20482838. Pang2010.
- ^ Reich D (29 March 2018). Who We Are and How We Got Here: Ancient DNA and the new science of the human past. Oxford University Press. p. xxiv. ISBN 978-0-19-255438-3.
- ^ Livingstone F (Summer 1962). "On the Non-Existence of Human Races" (PDF). Chicago Journals. Archived from the original (PDF) on 25 May 2021. Retrieved 29 April 2019.
- ^ Honnay O (2013), "Genetic Drift", Brenner's Encyclopedia of Genetics, Elsevier, pp. 251–253, doi:10.1016/b978-0-12-374984-0.00616-1, ISBN 978-0-08-096156-9
- ^ Martin C, Zhang Y (June 2007). "Mechanisms of epigenetic inheritance". Current Opinion in Cell Biology. 19 (3): 266–272. doi:10.1016/j.ceb.2007.04.002. PMID 17466502.
- ^ a b c d Jorde LB, Wooding SP (November 2004). "Genetic variation, classification and 'race'". Nature Genetics. Supplemental. 36 (11). Nature Portfolio: 28–33. doi:10.1038/ng1435. ISSN 1546-1718. OCLC 8091998144. PMID 15508000. S2CID 1500915. Jorde2004. Archived from the original on 22 June 2024. Retrieved 27 June 2024.
- ^ Auton A, Brooks LD, Durbin RM, Garrison EP, et al. (October 2015). "A global reference for human genetic variation". Nature. 526 (7571): 68–74. Bibcode:2015Natur.526...68T. doi:10.1038/nature15393. PMC 4750478. PMID 26432245.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ Cavalli-Sforza LL, Menozzi P, Piazza A (1994). The History and Geography of Human Genes. Princeton: Princeton University Press. ISBN 978-0-691-08750-4.
- Mark Ridley (20 August 2000). "How Far From the Tree?". The New York Times (Review). Archived from the original on 17 March 2017. Retrieved 3 March 2017.
- ^ Norton HL, Quillen EE, Bigham AW, Pearson LN, et al. (9 July 2019). "Human races are not like dog breeds: refuting a racist analogy". Evolution: Education and Outreach. 12 (1): 17. doi:10.1186/s12052-019-0109-y. ISSN 1936-6434.
- ^ Templeton A (2008). "Human Races: A Genetic and Evolutionary Perspective". American Anthropologist. 100 (3): 633. doi:10.1525/aa.1998.100.3.632. Retrieved 19 July 2024.
- ^ Ostrander EA (6 February 2017). "Genetics and the Shape of Dogs". American Scientist. Retrieved 12 March 2024.
- ^ Bamshad M (2005). "Genetic Influenes on Health: Does Race Matter?". JAMA. 294 (8): 938. doi:10.1001/jama.294.8.937. PMID 16118384. Retrieved 18 July 2024.
- ^ Parker HG, Kim LV, Sutter NB, Carlson S, et al. (21 May 2004). "Genetic Structure of the Purebred Domestic Dog". Science. 304 (5674): 1160–1164. Bibcode:2004Sci...304.1160P. doi:10.1126/science.1097406. ISSN 0036-8075. PMID 15155949. Retrieved 18 July 2024.
- ^ Orsucci A (1998). "Ariani, indogermani, stirpi mediterranee: aspetti del dibattito sulle razze europee (1870–1914)". Cromohs (in Italian). Archived from the original on 18 December 2012.
- ^ Angier N (22 August 2000). "Do Races Differ? Not Really, DNA Shows". The New York Times. Archived from the original on 30 April 2021. Retrieved 3 September 2011.
- ^ Owens K, King MC (15 October 1999). "Genomic Views of Human History". Science. 286 (5439): 451–453. doi:10.1126/science.286.5439.451. ISSN 0036-8075. PMID 10521333.
Variation in other traits popularly used to identify 'races' is likely to be due to similarly straightforward mechanisms, involving limited numbers of genes with very specific physiological effects.
- ^ Ezkurdia I, Juan D, Rodriguez JM, Frankish A, et al. (November 2014). "Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes". Human Molecular Genetics. 23 (22): 5866–5878. doi:10.1093/hmg/ddu309. PMC 4204768. PMID 24939910.
- ^ Sturm RA, Duffy DL (2012). "Human pigmentation genes under environmental selection". Genome Biology. 13 (9): 248. doi:10.1186/gb-2012-13-9-248. ISSN 1474-760X. PMC 3491390. PMID 23110848.
- ^ White D, Rabago-Smith M (January 2011). "Genotype-phenotype associations and human eye color". Journal of Human Genetics. 56 (1): 5–7. doi:10.1038/jhg.2010.126. PMID 20944644.
- ^ Sykes B (2001). "From Blood Groups to Genes". The seven daughters of Eve. New York: Norton. pp. 32–51. ISBN 978-0-393-02018-2.
- ^ Blancher A, Klein J, Socha WW (2012). Molecular biology and evolution of blood group and MHC antigens in primates. Springer Science & Business Media. ISBN 978-3-642-59086-3.
- ^ Ségurel L, Thompson EE, Flutre T, Lovstad J, et al. (6 November 2012). "The ABO blood group is a trans-species polymorphism in primates". Proceedings of the National Academy of Sciences. 109 (45): 18493–18498. arXiv:1208.4613. Bibcode:2012PNAS..10918493S. doi:10.1073/pnas.1210603109. ISSN 0027-8424. PMC 3494955. PMID 23091028.
- ^ Lewontin R (1972). "The Apportionment of Human Diversity". In Theodosius Dobzhansky, Max K. Hecht, William C. Steere (eds.). Evolutionary Biology. Vol. 6. pp. 381–398. doi:10.1007/978-1-4684-9063-3_14. ISBN 978-1-4684-9065-7. S2CID 21095796.
- ^ Risch N, Burchard E, Ziv E, Tang H (2002). "Categorization of humans in biomedical research: genes, race and disease". Genome Biology. 3 (7): comment2007.1. doi:10.1186/gb-2002-3-7-comment2007. ISSN 1465-6906. PMC 139378. PMID 12184798.
- ^ Templeton AR (2003). "Human Races in the Context of Recent Human Evolution: A Molecular Genetic Perspective". In Goodman AH, Heath D, Lindee MS (eds.). Genetic nature/culture: anthropology and science beyond the two-culture divide. Berkeley: University of California Press. pp. 234–257. ISBN 978-0-520-23792-6. Archived from the original on 9 November 2014. Retrieved 23 September 2014.
- ^ Ossorio P, Duster T (January 2005). "Race and genetics: controversies in biomedical, behavioral, and forensic sciences". The American Psychologist. 60 (1): 115–128. doi:10.1037/0003-066X.60.1.115. PMID 15641926.
- ^ Lewontin RC (2005). "Confusions About Human Races". Race and Genomics. Social Sciences Research Council. Archived from the original on 4 May 2013. Retrieved 28 December 2006.
- ^ a b Long JC, Kittles RA (2009). "Human Genetic Diversity and the Nonexistence of Biological Races". Human Biology. 81 (5): 777–798. doi:10.3378/027.081.0621. ISSN 1534-6617. PMID 20504196. S2CID 30709062. Archived from the original on 13 March 2020. Retrieved 13 January 2016.
- ^ Edwards AW (August 2003). "Human genetic diversity: Lewontin's fallacy". BioEssays. 25 (8): 798–801. doi:10.1002/bies.10315. PMID 12879450.
- ^ Winther RG (2018). Phylogenetic Inference, Selection Theory, and History of Science: Selected Papers of A. W. F. Edwards with Commentaries. Cambridge, U.K.: Cambridge University Press. ISBN 9781107111721. Archived from the original on 15 August 2019. Retrieved 13 December 2018.
- ^ Edwards A (2003). Human genetic diversity: Lewontin's fallacy, BioEssays. pp. 798–801.
- ^ Sesardic N (2010). Race: a social destruction of a biological concept. Biology and Philosophy. pp. 143–162.
- ^ Winther R (2018). "The Genetic Reification of "Race"? A Story of Two Mathematical Methods". In R.G. Winther (ed.). Phylogenetic Inference, Selection Theory, and History of Science: Selected Papers of AWF Edwards with Commentaries. Cambridge University Press. pp. 489, 488–508. ISBN 9781107111721. Archived from the original on 15 August 2019. Retrieved 13 December 2018.
- ^ Wohns AW, Wong Y, Jeffery B, Akbari A, et al. (15 April 2021). "A unified genealogy of modern and ancient genomes". bioRxiv 10.1101/2021.02.16.431497.
- ^ Witherspoon DJ, Wooding S, Rogers AR, Marchani EE, et al. (2007). "Genetic Similarities Within and Between Human Populations". Genetics. 176 (1): 351–359. doi:10.1534/genetics.106.067355. ISSN 0016-6731. PMC 1893020. PMID 17339205.
- ^ a b c d e Tang H, Quertermous T, Rodriguez B, et al. (February 2005). "Genetic Structure, Self-Identified Race/Ethnicity, and Confounding in Case-Control Association Studies". American Journal of Human Genetics. 76 (2): 268–75. doi:10.1086/427888. PMC 1196372. PMID 15625622.
- ^ Lewontin RC. "Confusions About Human Races". Archived from the original on 4 May 2013. Retrieved 9 January 2007.
- ^ Kittles RA, Weiss KM (2003). "Race, ancestry, and genes: implications for defining disease risk". Annual Review of Genomics and Human Genetics. 4: 33–67. doi:10.1146/annurev.genom.4.070802.110356. PMID 14527296.
- ^ a b c Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, et al. (December 2005). "Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure". PLOS Genetics. 1 (6): e70. doi:10.1371/journal.pgen.0010070. PMC 1310579. PMID 16355252.
- ^ Li JZ, Absher DM, Tang H, Southwick AM, et al. (2008). "Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation". Science. 319 (5866): 1100–1104. Bibcode:2008Sci...319.1100L. doi:10.1126/science.1153717. PMID 18292342. S2CID 53541133.
- ^ Jakobsson M, Scholz SW, Scheet P, Gibbs JR, et al. (2008). "Genotype, haplotype and copy-number variation in worldwide human populations" (PDF). Nature. 451 (7181): 998–1003. Bibcode:2008Natur.451..998J. doi:10.1038/nature06742. hdl:2027.42/62552. PMID 18288195. S2CID 11074384.
- ^ Xing J, Watkins WS, Witherspoon DJ, Zhang Y, et al. (2009). "Fine-scaled human genetic structure revealed by SNP microarrays". Genome Research. 19 (5): 815–825. doi:10.1101/gr.085589.108. PMC 2675970. PMID 19411602.
- ^ López Herráez D, Bauchet M, Tang K, Theunert C, et al. (2009). Hawks J (ed.). "Genetic Variation and Recent Positive Selection in Worldwide Human Populations: Evidence from Nearly 1 Million SNPs". PLOS ONE. 4 (11): e7888. Bibcode:2009PLoSO...4.7888L. doi:10.1371/journal.pone.0007888. PMC 2775638. PMID 19924308.
- ^ Biddanda A, Rice DP, Novembre J (2020). "A variant-centric perspective on geographic patterns of human allele frequency variation". eLife. 9. doi:10.7554/eLife.60107. PMC 7755386. PMID 33350384.
- ^ Ramachandran S, Deshpande O, Roseman CC, Rosenberg NA, et al. (November 2005). "Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa". Proceedings of the National Academy of Sciences. 102 (44): 15942–7. Bibcode:2005PNAS..10215942R. doi:10.1073/pnas.0507611102. PMC 1276087. PMID 16243969.
- ^ Harpending H (1 November 2002). "Kinship and Population Subdivision" (PDF). Population & Environment. 24 (2): 141–147. doi:10.1023/A:1020815420693. JSTOR 27503827. S2CID 15208802. Archived (PDF) from the original on 28 June 2017. Retrieved 22 August 2017.
- ^ a b Long, J. C., Kittles, R. A. (2009). "Human genetic diversity and the nonexistence of biological races". Human Biology. 81 (5/6): 777–798. doi:10.3378/027.081.0621. PMID 20504196. S2CID 30709062.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ a b Mountain, J. L., Risch, N. (2004). "Assessing genetic contributions to phenotypic differences among 'racial' and 'ethnic' groups". Nature Genetics. 36 (11 Suppl): S48–S53. doi:10.1038/ng1456. PMID 15508003.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ a b Pearse DE, Crandall KA (2004). "Beyond FST: analysis of population genetic data for conservation". Conservation Genetics. 5 (5): 585–602. Bibcode:2004ConG....5..585P. doi:10.1007/s10592-003-1863-4. S2CID 22068080.
- ^ Hunley KL, Cabana GS, Long JC (1 December 2015). "The apportionment of human diversity revisited". American Journal of Physical Anthropology. 160 (4): 561–569. doi:10.1002/ajpa.22899. ISSN 1096-8644. PMID 26619959.
- ^ Brace CL (2005). "Race" is a Four-letter Word: The Genesis of the Concept. Oxford: Oxford University Press. ISBN 978-0-19-517351-2.
- ^ Graves JL (2001). The Emperor's New Clothes: Biological Theories of Race at the Millennium. Rutgers University Press. ISBN 9780813528472.
- ^ Weiss KM, Fullerton SM (2005). "Racing around, getting nowhere". Evolutionary Anthropology: Issues, News, and Reviews. 14 (5): 165–169. doi:10.1002/evan.20079. ISSN 1060-1538. S2CID 84927946.
- ^ Kaplan JM (17 January 2011). "'Race': What Biology Can Tell Us about a Social Construct". Encyclopedia of Life Sciences (ELS). doi:10.1002/9780470015902.a0005857. ISBN 978-0470016176. Retrieved 23 September 2014.
- ^ a b c d Cavalli-Sforza LL (1997). "Genes, peoples, and languages". Proceedings of the National Academy of Sciences. 94 (15): 7719–7724. Bibcode:1997PNAS...94.7719C. doi:10.1073/pnas.94.15.7719. PMC 33682.
- ^ Paschou P, Lewis J, Javed A, Drineas P (2010). "Ancestry informative markers for fine-scale individual assignment to worldwide populations". J Med Genet. 47 (12): 835–847. doi:10.1136/jmg.2010.078212. PMID 20921023. S2CID 6432430. Archived from the original on 5 November 2018. Retrieved 4 November 2018.
- ^ Pena SD, Di Pietro G, Fuchshuber-Moraes M, Genro JP, et al. (2011). Harpending H (ed.). "The Genomic Ancestry of Individuals from Different Geographical Regions of Brazil is More Uniform Than Expected". PLoS ONE. 6 (2): e17063. Bibcode:2011PLoSO...617063P. doi:10.1371/journal.pone.0017063. PMC 3040205. PMID 21359226.
- ^ Parra FC (2002). "Color and genomic ancestry in Brazilians". Proceedings of the National Academy of Sciences. 100 (1): 177–182. Bibcode:2003PNAS..100..177P. doi:10.1073/pnas.0126614100. PMC 140919. PMID 12509516.
- ^ Lima-Costa MF, Rodrigues LC, Barreto ML, Gouveia M, et al. (2015). "Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative)". Scientific Reports. 5: 9812. Bibcode:2015NatSR...5E9812.. doi:10.1038/srep09812. PMC 5386196. PMID 25913126.
- ^ Frank R. "Back with a Vengeance: the Reemergence of a Biological Conceptualization of Race in Research on Race/Ethnic Disparities in Health". Archived from the original on 1 December 2008.
- ^ Bolnick DA (2008). "Individual Ancestry Inference and the Reification of Race as a Biological Phenomenon". In Koenig BA, Richardson SS, Lee SS (eds.). Revisiting race in a genomic age. Rutgers University Press. ISBN 978-0-8135-4324-6.
- ^ Patterson N, Price AL, Reich D (2006). "Population Structure and Eigenanalysis". PLOS Genetics. 2 (12): e190. doi:10.1371/journal.pgen.0020190. PMC 1713260. PMID 17194218.
- ^ Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, et al. (2005). "Clines, Clusters, and the Effect of Study Design on the Inference of Human Population Structure". PLOS Genet. 1 (6): e70. doi:10.1371/journal.pgen.0010070. PMC 1310579. PMID 16355252.
{{cite journal}}: CS1 maint: overridden setting (link) - ^ a b Duster T (March 2015). "A post-genomic surprise. The molecular reinscription of race in science, law and medicine". The British Journal of Sociology. 66 (1): 1–27. doi:10.1111/1468-4446.12118. ISSN 0007-1315. PMID 25789799.
- ^ Fullwiley, D. (2008). "The Biologistical Construction of Race: 'Admixture' Technology and the New Genetic Medicine". Social Studies of Science, 38(5), 695–735. doi:10.1177/0306312708090796
- ^ Risch N (July 2005). "The whole side of it--an interview with Neil Risch by Jane Gitschier". PLOS Genetics. 1 (1): e14. doi:10.1371/journal.pgen.0010014. PMC 1183530. PMID 17411332.
- ^ "Malaria and the Red Cell". Harvard University. 2002. Archived from the original on 27 November 2011.
- ^ Högenauer C, Santa Ana CA, Porter JL, et al. (December 2000). "Active intestinal chloride secretion in human carriers of cystic fibrosis mutations: an evaluation of the hypothesis that heterozygotes have subnormal active intestinal chloride secretion". Am. J. Hum. Genet. 67 (6): 1422–1427. doi:10.1086/316911. PMC 1287919. PMID 11055897.
- ^ Schwartz RS (2001). "Racial Differences in the Response to Drugs — Pointers to Genetic Differences". New England Journal of Medicine. 344 (18): 1393–1396. doi:10.1056/NEJM200105033441810. PMID 11333999. Archived from the original on 1 September 2003. Retrieved 28 October 2009.
- ^ Bloche GM (2004). "Race-Based Therapeutics". New England Journal of Medicine. 351 (20): 2035–2037. doi:10.1056/nejmp048271. PMID 15533852.
- ^ Drug information for the drug Crestor Archived 2009-09-26 at the Wayback Machine. Warnings for this drug state, "People of Asian descent may absorb rosuvastatin at a higher rate than other people. Make sure your doctor knows if you are Asian. You may need a lower than normal starting dose."
- ^ Risch N, Burchard E, Ziv E, Tang H (2002). "Categorization of humans in biomedical research: genes, race and disease". Genome Biol. 3 (7): 1–12. doi:10.1186/gb-2002-3-7-comment2007. PMC 139378. PMID 12184798.
- ^ "Census, Race and Science". Nature Genetics. 24 (2): 97–98. 2000. doi:10.1038/72884. PMID 10655044.
- ^ "Geneticists curb use of 'race'". Science. 374 (6572): 1177. 3 December 2021.
- ^ "Revelations". Shut Up and Listen. Palgrave Macmillan. 2011. doi:10.1057/9780230362987.0004. ISBN 978-0-230-36298-7. Retrieved 28 January 2021..
- ^ Spencer Q (2015). "Philosophy of race meets population genetics". Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 49. doi:10.1016/j.shpsc.2015.04.003. PMID 25963045.
- ^ Spencer Q (2015). "Philosophy of race meets population genetics". Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 51. doi:10.1016/j.shpsc.2015.04.003. PMID 25963045.
- ^ Spencer Q (2015). "Philosophy of race meets population genetics". Studies in History and Philosophy of Biological and Biomedical Sciences. 52: 46–47. doi:10.1016/j.shpsc.2015.04.003. PMID 25963045.
- ^ a b Zach N (2003). Philosophy of Science and Race. Taylor & Francis. ISBN 9781134728022.
- ^ Sundstrom R (2002). "Race as a human kind". Philosophy & Social Criticism. 28: 91–115. doi:10.1177/0191453702028001592. S2CID 145381236.
- ^ Witherspoon DJ, Wooding S, Rogers AR, et al. (May 2007). "Genetic Similarities Within and Between Human Populations". Genetics. 176 (1): 351–9. doi:10.1534/genetics.106.067355. PMC 1893020. PMID 17339205.
- ^ Witherspoon DJ, Wooding S, Rogers AR, et al. (May 2007). "Genetic Similarities Within and Between Human Populations". Genetics. 176 (1): 358. doi:10.1534/genetics.106.067355. PMC 1893020. PMID 17339205.
- ^ Sauer NJ (January 1992). "Forensic anthropology and the concept of race: if races don't exist, why are forensic anthropologists so good at identifying them?". Social Science & Medicine. 34 (2): 107–111. doi:10.1016/0277-9536(92)90086-6. PMID 1738862.
- ^ Duster T (2015). "A post-genomic surprise. The molecular reinscription of race in science, law and medicine". The British Journal of Sociology. 66 (1): 1–27. doi:10.1111/1468-4446.12118. PMID 25789799.
- ^ Hunt LM, Megyesi M (Fall 2007). "The ambiguous meanings of the racial/ethnic categories routinely used in human genetics research". Social Science and Medicine. 66 (2): 349–361. doi:10.1016/j.socscimed.2007.08.034. PMC 2213883. PMID 17959289 – via Science Direct Assets.
Further reading
[edit]- Glasgow J (2009). A Theory of Race. New York: Routledge. ISBN 9780415990721.
- Helms JE, Jernigan M, Mascher J (January 2005). "The meaning of race in psychology and how to change it: a methodological perspective" (PDF). The American Psychologist. 60 (1): 27–36. doi:10.1037/0003-066X.60.1.27. PMID 15641919. S2CID 1676488. Archived from the original (PDF) on 26 February 2019.
- Keita SO, Kittles RA, Royal CD, et al. (November 2004). "Conceptualizing human variation". Nature Genetics. 36 (11 Suppl): S17–20. doi:10.1038/ng1455. PMID 15507998.
- Koenig BA, Lee SS, Richardson SS, eds. (2008). Revisiting Race in a Genomic Age. New Brunswick (NJ): Rutgers University Press. ISBN 978-0-8135-4324-6. This review of current research includes chapters by Jonathan Marks, John Dupré, Sally Haslanger, Deborah A. Bolnick, Marcus W. Feldman, Richard C. Lewontin, Sarah K. Tate, David B. Goldstein, Jonathan Kahn, Duana Fullwiley, Molly J. Dingel, Barbara A. Koenig, Mark D. Shriver, Rick A. Kittles, Henry T. Greely, Kimberly Tallbear, Alondra Nelson, Pamela Sankar, Sally Lehrman, Jenny Reardon, Jacqueline Stevens, and Sandra Soo-Jin Lee.
- Lieberman L, Kirk RC, Corcoran M (2003). "The Decline of Race in American Physical Anthropology" (PDF). Przegląd Antropologiczny – Anthropological Review. 66: 3–21. ISSN 0033-2003. Archived from the original (PDF) on 8 June 2011. Retrieved 12 September 2010.
- Long JC, Kittles RA (August 2003). "Human genetic diversity and the nonexistence of biological races". Human Biology. 75 (4): 449–71. doi:10.1353/hub.2003.0058. PMID 14655871. S2CID 26108602.
- Miththapala S, Seidensticker J, O'Brien SJ (1996). "Phylogeographic Subspecies Recognition in Leopards (Panthera pardus): Molecular Genetic Variation". Conservation Biology. 10 (4): 1115–1132. Bibcode:1996ConBi..10.1115M. doi:10.1046/j.1523-1739.1996.10041115.x.
- Ossorio P, Duster T (January 2005). "Race and genetics: controversies in biomedical, behavioral, and forensic sciences". The American Psychologist. 60 (1): 115–28. doi:10.1037/0003-066X.60.1.115. PMID 15641926.
- Parra EJ, Kittles RA, Shriver MD (November 2004). "Implications of correlations between skin color and genetic ancestry for biomedical research". Nature Genetics. 36 (11 Suppl): S54–60. doi:10.1038/ng1440. PMID 15508005.
- Sawyer SL, Mukherjee N, Pakstis AJ, et al. (May 2005). "Linkage disequilibrium patterns vary substantially among populations". European Journal of Human Genetics. 13 (5): 677–86. doi:10.1038/sj.ejhg.5201368. PMID 15657612.
- Rohde DL, Olson S, Chang JT (September 2004). "Modelling the recent common ancestry of all living humans". Nature. 431 (7008): 562–6. Bibcode:2004Natur.431..562R. CiteSeerX 10.1.1.78.8467. doi:10.1038/nature02842. PMID 15457259. S2CID 3563900.
- Serre D, Pääbo S (September 2004). "Evidence for Gradients of Human Genetic Diversity Within and Among Continents". Genome Research. 14 (9): 1679–85. doi:10.1101/gr.2529604. PMC 515312. PMID 15342553.
- Smedley A, Smedley BD (January 2005). "Race as biology is fiction, racism as a social problem is real: Anthropological and historical perspectives on the social construction of race". The American Psychologist. 60 (1): 16–26. CiteSeerX 10.1.1.568.4548. doi:10.1037/0003-066X.60.1.16. PMID 15641918.
