Pilin: Difference between revisions
No edit summary Tags: Mobile edit Mobile web edit |
Citation bot (talk | contribs) Alter: volume, url. URLs might have been anonymized. Add: issue, doi-access. | Use this bot. Report bugs. | #UCB_CommandLine |
||
| (39 intermediate revisions by 11 users not shown) | |||
| Line 1: | Line 1: | ||
{{missing information|article|explanation of convergent evolution and a list of classes|date=December 2020}} |
|||
{{split|type IV pilin|Saf pilin|date=December 2020}} |
|||
| ⚫ | '''Pilin''' refers to a class of fibrous [[protein]]s that are found in [[pilus]] structures in [[bacteria]]. These structures can be used for the exchange of [[gene]]tic material, or as a [[cell adhesion]] mechanism. Although not all bacteria have pili or fimbriae, bacterial [[pathogen]]s often use their fimbriae to attach to host cells. In [[Gram-negative]] bacteria, where pili are more common, individual pilin molecules are linked by [[noncovalent]] [[protein-protein interaction]]s, while [[Gram-positive]] bacteria often have [[polymer]]ized LPXTG pilin.<ref name="pmid16778837">{{cite journal |vauthors=Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G |title=Pili in gram-positive pathogens |journal=Nat. Rev. Microbiol. |volume=4 |issue=7 |pages=509–19 |year=2006 |pmid=16778837 |doi=10.1038/nrmicro1443 |s2cid=6369483 |doi-access=free }}</ref> |
||
== Type IV pilin == |
|||
{{see also|Type IV filament|Type IV pili}} |
|||
{{Pfam_box |
{{Pfam_box |
||
| Symbol = Pili |
| Symbol = Pili |
||
| Name = Pilin |
| Name = Pilin in Type IV pili |
||
| image = Pilin-2pil.png |
| image = Pilin-2pil.png |
||
| width = 200 px |
| width = 200 px |
||
| caption = Pilin protein from ''[[Neisseria gonorrhoeae]]'', a [[parasite|parasitic]] bacterium that requires functional pili for pathogenesis. |
| caption = Pilin protein from ''[[Neisseria gonorrhoeae]]'', a [[parasite|parasitic]] bacterium that requires functional pili for [[pathogenesis]]. |
||
| Pfam= PF00114 |
| Pfam= PF00114 |
||
| InterPro= IPR001082 |
| InterPro= IPR001082 |
||
| Line 11: | Line 18: | ||
| SCOP = 1paj |
| SCOP = 1paj |
||
| TCDB = |
| TCDB = |
||
| OPM family= |
| OPM family= 68 |
||
| OPM protein= 2hil |
| OPM protein= 2hil |
||
| PDB= |
| PDB= |
||
}} |
}} |
||
| ⚫ | Type IV pilin proteins are [[alpha+beta protein fold|α+β]] proteins characterized by a very long [[N-terminus|N-terminal]] [[alpha helix]]. The assembly of these pili relies on interactions between the N-terminal helices of the individual monomers. The pilus structure sequesters the helices in the center of the fiber lining a central pore, while antiparallel [[beta sheet]]s occupy the exterior of the fiber.<ref name="pmid9224887">{{cite journal |vauthors=Forest KT, Tainer JA |title=Type-4 pilus-structure: outside to inside and top to bottom--a minireview |journal=Gene |volume=192 |issue=1 |pages=165–9 |year=1997 |pmid=9224887 |doi= 10.1016/s0378-1119(97)00008-5}}</ref> |
||
| ⚫ | '''Pilin''' refers to a class of fibrous [[protein]]s that are found in [[pilus]] structures in [[bacteria]]. |
||
=== Role of ComP pilin in bacterial transformation === |
|||
| ⚫ | |||
| ⚫ | [[Transformation (genetics)|Genetic transformation]] is the process by which a recipient bacterial cell takes up DNA from a neighboring cell and integrates this DNA into the recipient’s genome by [[homologous recombination]]. In ''[[Neisseria meningitidis]]'', DNA transformation requires the presence of short [[DNA uptake sequence]]s (DUSs) which are 9-10mers residing in [[coding region]]s of the donor DNA. Specific recognition of DUSs is mediated by a type IV pilin, ComP.<ref name="pmid24385921">{{cite journal |vauthors=Berry JL, Cehovin A, McDowell MA, Lea SM, Pelicic V |title=Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species |journal=PLOS Genet. |volume=9 |issue=12 |pages=e1004014 |year=2013 |pmid=24385921 |pmc=3868556 |doi=10.1371/journal.pgen.1004014 |doi-access=free }}</ref><ref name="pmid23386723">{{cite journal |vauthors=Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V |title=Specific DNA recognition mediated by a type IV pilin |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=110 |issue=8 |pages=3065–70 |year=2013 |pmid=23386723 |pmc=3581936 |doi=10.1073/pnas.1218832110 |bibcode=2013PNAS..110.3065C |doi-access=free }}</ref> Menningococcal type IV pili bind DNA through the minor pilin ComP via an electropositive stripe that is predicted to be exposed on the filament's surface. ComP displays an exquisite binding preference for selective DUSs. The distribution of DUSs within the ''N. meningitidis'' genome favors certain genes, suggesting that there is a bias for genes involved in genomic maintenance and repair.<ref name="pmid14960717">{{cite journal |vauthors=Davidsen T, Rødland EA, Lagesen K, Seeberg E, Rognes T, Tønjum T |title=Biased distribution of DNA uptake sequences towards genome maintenance genes |journal=Nucleic Acids Res. |volume=32 |issue=3 |pages=1050–8 |year=2004 |pmid=14960717 |pmc=373393 |doi=10.1093/nar/gkh255 }}</ref><ref name="pmid19464092">{{cite journal |vauthors=Caugant DA, Maiden MC |title=Meningococcal carriage and disease--population biology and evolution |journal=Vaccine |volume=27 |pages=B64–70 |year=2009 |issue=Suppl 2 |pmid=19464092 |pmc=2719693 |doi=10.1016/j.vaccine.2009.04.061 }}</ref> |
||
| ⚫ | |||
| ⚫ | Pili in [[Gram-positive bacteria]] contain spontaneously formed [[isopeptide bond]]s. These bonds provide enhanced mechanical<ref name="pmid20139067">{{cite journal |vauthors=Alegre-Cebollada J, Badilla CL, Fernández JM |title=Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes |journal=J. Biol. Chem. |volume=285 |issue=15 |pages=11235–11242 |year=2010 |pmid=20139067 |doi=10.1074/jbc.M110.102962 |pmc=2857001}}</ref> and proteolytic<ref>{{cite journal | vauthors = Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN | year = 2007 | title = Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure |
||
== |
== Chaperone-usher pilin == |
||
{{main|chaperone-usher fimbriae}} |
|||
The Cup family is known for its use of a [[chaperone (protein)|chaperone]] and at least an [[Fimbrial usher protein|usher]]. They exhibit an Ig fold.<ref name="Verger D et al2007">{{cite journal |vauthors=Verger D, etal | title=Crystal structure of the P-pilus rob subunit PapA | journal=PLOS ONE | year=2007 | doi=10.1371/journal.ppat.0030073 | volume=3 | issue=5 | pages=e73| pmc=1868955 | pmid=17511517 | doi-access=free }}</ref> |
|||
| ⚫ | [[Transformation (genetics)|Genetic transformation]] is the process by which a recipient bacterial cell takes up DNA from a neighboring cell and integrates this DNA into the recipient’s genome by [[homologous recombination]]. In ''[[Neisseria |
||
===Saf, N-terminal extension=== |
|||
{{Infobox protein family |
|||
| Symbol = Saf-Nte_pilin |
|||
| Name = Saf-Nte_pilin |
|||
| image = PDB 2co1 EBI.jpg |
|||
| width = |
|||
| caption = ''Salmonella enterica'' SafA pilin in complex with a 19-residue SafA Nte peptide (f17a mutant) |
|||
| Pfam = PF09460 |
|||
| Pfam_clan = |
|||
| InterPro = IPR018569 |
|||
| SMART = |
|||
| PROSITE = |
|||
| MEROPS = |
|||
| SCOP = |
|||
| TCDB = |
|||
| OPM family = |
|||
| OPM protein = |
|||
| CAZy = |
|||
| CDD = |
|||
}} |
|||
The '''Saf pilin N-terminal extension''' [[protein domain]] helps the pili to form, via a complex mechanism named the [[Chaperone (protein)|chaperone]]/[[Fimbrial usher protein|usher]] pathway. It is found in all c-u pilins.<ref name=walkman>{{cite journal |last1=Waksman |first1=G |last2=Hultgren |first2=SJ |title=Structural biology of the chaperone-usher pathway of pilus biogenesis. |journal=Nature Reviews. Microbiology |date=November 2009 |volume=7 |issue=11 |pages=765–74 |doi=10.1038/nrmicro2220 |pmid=19820722 |pmc=3790644 |doi-access=free}}</ref> |
|||
This protein domain is very important for such bacteria, as without pili formation, they could not infect the host. Saf is a ''Salmonella'' [[operon]] containing a c-u pilus system.<ref name=walkman/> |
|||
===Function=== |
|||
This protein domain, has an important function in forming pili. These are virulence factors crucial for [[cell adhesion]] to the host and [[biofilm]] formation with successful infection.<ref name="pmid18448124">{{cite journal | vauthors = Salih O, Remaut H, Waksman G, Orlova EV | title = Structural analysis of the Saf pilus by electron microscopy and image processing | journal = Journal of Molecular Biology | volume = 379 | issue = 1 | pages = 174–87 | date = May 2008 | pmid = 18448124 | doi = 10.1016/j.jmb.2008.03.056 | url = https://pubmed.ncbi.nlm.nih.gov/18448124 }}</ref> |
|||
=== Structure === |
|||
This [[protein domain]] consists of the adjacent Saf-Nte and Saf-pilin [[polymer|chain]]s of the pilus-forming [[Protein complex|complex]]. They are Chaperone/usher (CU) pili, and have an [[N-terminal]] extension (Nte) of around 10-20 [[amino acids]]. Salmonella Saf pili, which are assembled by FGl chaperones. The structure has been well conserved, as they contain a set of alternating [[hydrophobic]] [[amino acid|residues]] that form an essential part of the subunit–subunit interaction.<ref name="pmid19820722">{{cite journal | vauthors = Waksman G, Hultgren SJ | title = Structural biology of the chaperone-usher pathway of pilus biogenesis | journal = Nature Reviews. Microbiology | volume = 7 | issue = 11 | pages = 765–74 | date = November 2009 | pmid = 19820722 | pmc = 3790644| doi = 10.1038/nrmicro2220 }}</ref> |
|||
===Mechanism=== |
|||
The mechanism for the assembly reaction is termed donor strand exchange '''DSE''' which |
|||
[[Pilus]] assembly in Gram-negative [[bacteria]] involves a Donor-strand exchange [[Nuclear receptor#Mechanism of action|mechanism]] between the C- and the N-termini of this domain. The C-terminal subunit forms an incomplete Ig-fold which is then complemented by the 10-18 [[Residue (chemistry)|residue]] N terminus of another. |
|||
The N terminus [[sequence (biology)|sequence]]s contain a [[protein motif|motif]] of alternating [[hydrophobic]] [[amino acid|residue]]s that occupy the P2 to P5 [[Binding (molecular)|binding]] pockets in the groove of the first pilus subunit.<ref name="pmid16793551">{{cite journal | vauthors = Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G | title = Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism | journal = Molecular Cell | volume = 22 | issue = 6 | pages = 831–42 | date = June 2006 | pmid = 16793551 | doi = 10.1016/j.molcel.2006.05.033 | doi-access = free }}</ref> |
|||
== LPXTG pilin == |
|||
<!-- Pfam PF00746 --> |
|||
LPXTG pilin is common in [[gram-positive]] cocci. They are named for a C-terminal motif used by the [[sortase]].<ref name="pmid16778837"/> There is also a [[LPXTGase]]. |
|||
| ⚫ | |||
| ⚫ | LPXTG Pili in [[Gram-positive bacteria]] contain spontaneously formed [[isopeptide bond]]s. These bonds provide enhanced mechanical<ref name="pmid20139067">{{cite journal |vauthors=Alegre-Cebollada J, Badilla CL, Fernández JM |title=Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes |journal=J. Biol. Chem. |volume=285 |issue=15 |pages=11235–11242 |year=2010 |pmid=20139067 |doi=10.1074/jbc.M110.102962 |pmc=2857001|doi-access=free }}</ref> and proteolytic<ref>{{cite journal | vauthors = Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN | year = 2007 | title = Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure | journal = Science | volume = 318 | issue = 5856| pages = 1625–1628 | doi=10.1126/science.1145806 | pmid=18063798| bibcode = 2007Sci...318.1625K | s2cid = 5627277 }}</ref> stability to the pilin protein. Recently, the pilin protein from ''[[Streptococcus pyogenes]]'' has been split into two fragments to develop a new molecular tool called the [[isopeptag]].<ref name="pmid20235501">{{cite journal |vauthors=Zakeri B, Howarth M |title=Spontaneous intermolecular amide bond formation between side chains for irreversible peptide targeting |journal=J. Am. Chem. Soc. |volume=132 |issue=13 |pages=4526–7 |year=2010 |pmid=20235501 |doi=10.1021/ja910795a |citeseerx=10.1.1.706.4839 }}</ref> The [[isopeptag]] is a short peptide that can be attached to a protein of interest and can bind its binding partner through a spontaneously formed [[isopeptide bond]]. This new peptide tag can allow scientists to target and isolate their proteins of interest through a permanent [[covalent bond]]. |
||
==See also== |
==See also== |
||
* |
* {{Annotated link|Prepilin peptidase}} |
||
==References== |
==References== |
||
{{Reflist}} |
{{Reflist}} |
||
{{InterPro content|IPR018569}} |
|||
== Further reading == |
|||
* {{cite journal |last1=Khare |first1=Baldeep |last2=V. L. Narayana |first2=Sthanam |title=Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly: Sortases and Implications for Assembly |journal=Protein Science |date=August 2017 |volume=26 |issue=8 |pages=1458–1473 |doi=10.1002/pro.3191|pmid=28493331 |pmc=5521585 |doi-access=free }} <!-- LPXTG stuff --> |
|||
[[Category:Bacterial proteins]] |
[[Category:Bacterial proteins]] |
||
Latest revision as of 19:23, 28 August 2023
This article is missing information about explanation of convergent evolution and a list of classes. (December 2020) |
It has been suggested that this article should be split into articles titled type IV pilin and Saf pilin. (discuss) (December 2020) |
Pilin refers to a class of fibrous proteins that are found in pilus structures in bacteria. These structures can be used for the exchange of genetic material, or as a cell adhesion mechanism. Although not all bacteria have pili or fimbriae, bacterial pathogens often use their fimbriae to attach to host cells. In Gram-negative bacteria, where pili are more common, individual pilin molecules are linked by noncovalent protein-protein interactions, while Gram-positive bacteria often have polymerized LPXTG pilin.[1]
Type IV pilin
[edit]| Pilin in Type IV pili | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Pilin protein from Neisseria gonorrhoeae, a parasitic bacterium that requires functional pili for pathogenesis. | |||||||||
| Identifiers | |||||||||
| Symbol | Pili | ||||||||
| Pfam | PF00114 | ||||||||
| InterPro | IPR001082 | ||||||||
| PROSITE | PDOC00342 | ||||||||
| SCOP2 | 1paj / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 68 | ||||||||
| OPM protein | 2hil | ||||||||
| |||||||||
Type IV pilin proteins are α+β proteins characterized by a very long N-terminal alpha helix. The assembly of these pili relies on interactions between the N-terminal helices of the individual monomers. The pilus structure sequesters the helices in the center of the fiber lining a central pore, while antiparallel beta sheets occupy the exterior of the fiber.[2]
Role of ComP pilin in bacterial transformation
[edit]Genetic transformation is the process by which a recipient bacterial cell takes up DNA from a neighboring cell and integrates this DNA into the recipient’s genome by homologous recombination. In Neisseria meningitidis, DNA transformation requires the presence of short DNA uptake sequences (DUSs) which are 9-10mers residing in coding regions of the donor DNA. Specific recognition of DUSs is mediated by a type IV pilin, ComP.[3][4] Menningococcal type IV pili bind DNA through the minor pilin ComP via an electropositive stripe that is predicted to be exposed on the filament's surface. ComP displays an exquisite binding preference for selective DUSs. The distribution of DUSs within the N. meningitidis genome favors certain genes, suggesting that there is a bias for genes involved in genomic maintenance and repair.[5][6]
Chaperone-usher pilin
[edit]The Cup family is known for its use of a chaperone and at least an usher. They exhibit an Ig fold.[7]
Saf, N-terminal extension
[edit]| Saf-Nte_pilin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
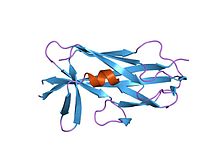 Salmonella enterica SafA pilin in complex with a 19-residue SafA Nte peptide (f17a mutant) | |||||||||
| Identifiers | |||||||||
| Symbol | Saf-Nte_pilin | ||||||||
| Pfam | PF09460 | ||||||||
| InterPro | IPR018569 | ||||||||
| |||||||||
The Saf pilin N-terminal extension protein domain helps the pili to form, via a complex mechanism named the chaperone/usher pathway. It is found in all c-u pilins.[8]
This protein domain is very important for such bacteria, as without pili formation, they could not infect the host. Saf is a Salmonella operon containing a c-u pilus system.[8]
Function
[edit]This protein domain, has an important function in forming pili. These are virulence factors crucial for cell adhesion to the host and biofilm formation with successful infection.[9]
Structure
[edit]This protein domain consists of the adjacent Saf-Nte and Saf-pilin chains of the pilus-forming complex. They are Chaperone/usher (CU) pili, and have an N-terminal extension (Nte) of around 10-20 amino acids. Salmonella Saf pili, which are assembled by FGl chaperones. The structure has been well conserved, as they contain a set of alternating hydrophobic residues that form an essential part of the subunit–subunit interaction.[10]
Mechanism
[edit]The mechanism for the assembly reaction is termed donor strand exchange DSE which Pilus assembly in Gram-negative bacteria involves a Donor-strand exchange mechanism between the C- and the N-termini of this domain. The C-terminal subunit forms an incomplete Ig-fold which is then complemented by the 10-18 residue N terminus of another.
The N terminus sequences contain a motif of alternating hydrophobic residues that occupy the P2 to P5 binding pockets in the groove of the first pilus subunit.[11]
LPXTG pilin
[edit]LPXTG pilin is common in gram-positive cocci. They are named for a C-terminal motif used by the sortase.[1] There is also a LPXTGase.
Development of molecular tools
[edit]LPXTG Pili in Gram-positive bacteria contain spontaneously formed isopeptide bonds. These bonds provide enhanced mechanical[12] and proteolytic[13] stability to the pilin protein. Recently, the pilin protein from Streptococcus pyogenes has been split into two fragments to develop a new molecular tool called the isopeptag.[14] The isopeptag is a short peptide that can be attached to a protein of interest and can bind its binding partner through a spontaneously formed isopeptide bond. This new peptide tag can allow scientists to target and isolate their proteins of interest through a permanent covalent bond.
See also
[edit]- Prepilin peptidase – Enzyme responsible for maturing type 4 pilins
References
[edit]- ^ a b Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G (2006). "Pili in gram-positive pathogens". Nat. Rev. Microbiol. 4 (7): 509–19. doi:10.1038/nrmicro1443. PMID 16778837. S2CID 6369483.
- ^ Forest KT, Tainer JA (1997). "Type-4 pilus-structure: outside to inside and top to bottom--a minireview". Gene. 192 (1): 165–9. doi:10.1016/s0378-1119(97)00008-5. PMID 9224887.
- ^ Berry JL, Cehovin A, McDowell MA, Lea SM, Pelicic V (2013). "Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species". PLOS Genet. 9 (12): e1004014. doi:10.1371/journal.pgen.1004014. PMC 3868556. PMID 24385921.
- ^ Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V (2013). "Specific DNA recognition mediated by a type IV pilin". Proc. Natl. Acad. Sci. U.S.A. 110 (8): 3065–70. Bibcode:2013PNAS..110.3065C. doi:10.1073/pnas.1218832110. PMC 3581936. PMID 23386723.
- ^ Davidsen T, Rødland EA, Lagesen K, Seeberg E, Rognes T, Tønjum T (2004). "Biased distribution of DNA uptake sequences towards genome maintenance genes". Nucleic Acids Res. 32 (3): 1050–8. doi:10.1093/nar/gkh255. PMC 373393. PMID 14960717.
- ^ Caugant DA, Maiden MC (2009). "Meningococcal carriage and disease--population biology and evolution". Vaccine. 27 (Suppl 2): B64–70. doi:10.1016/j.vaccine.2009.04.061. PMC 2719693. PMID 19464092.
- ^ Verger D, et al. (2007). "Crystal structure of the P-pilus rob subunit PapA". PLOS ONE. 3 (5): e73. doi:10.1371/journal.ppat.0030073. PMC 1868955. PMID 17511517.
- ^ a b Waksman, G; Hultgren, SJ (November 2009). "Structural biology of the chaperone-usher pathway of pilus biogenesis". Nature Reviews. Microbiology. 7 (11): 765–74. doi:10.1038/nrmicro2220. PMC 3790644. PMID 19820722.
- ^ Salih O, Remaut H, Waksman G, Orlova EV (May 2008). "Structural analysis of the Saf pilus by electron microscopy and image processing". Journal of Molecular Biology. 379 (1): 174–87. doi:10.1016/j.jmb.2008.03.056. PMID 18448124.
- ^ Waksman G, Hultgren SJ (November 2009). "Structural biology of the chaperone-usher pathway of pilus biogenesis". Nature Reviews. Microbiology. 7 (11): 765–74. doi:10.1038/nrmicro2220. PMC 3790644. PMID 19820722.
- ^ Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G (June 2006). "Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism". Molecular Cell. 22 (6): 831–42. doi:10.1016/j.molcel.2006.05.033. PMID 16793551.
- ^ Alegre-Cebollada J, Badilla CL, Fernández JM (2010). "Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes". J. Biol. Chem. 285 (15): 11235–11242. doi:10.1074/jbc.M110.102962. PMC 2857001. PMID 20139067.
- ^ Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN (2007). "Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure". Science. 318 (5856): 1625–1628. Bibcode:2007Sci...318.1625K. doi:10.1126/science.1145806. PMID 18063798. S2CID 5627277.
- ^ Zakeri B, Howarth M (2010). "Spontaneous intermolecular amide bond formation between side chains for irreversible peptide targeting". J. Am. Chem. Soc. 132 (13): 4526–7. CiteSeerX 10.1.1.706.4839. doi:10.1021/ja910795a. PMID 20235501.
Further reading
[edit]- Khare, Baldeep; V. L. Narayana, Sthanam (August 2017). "Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly: Sortases and Implications for Assembly". Protein Science. 26 (8): 1458–1473. doi:10.1002/pro.3191. PMC 5521585. PMID 28493331.
