Hepadnaviridae: Difference between revisions
m Protected "Hepadnaviridae": copyright violation - suspected anonymous educational project ([Edit=Require extended confirmed access] (expires 22:28, 20 June 2020 (UTC)) [Move=Require extended confirmed access] (expires 22:28, 20 June 2020 (UTC))) |
→top: correction |
||
| Line 16: | Line 16: | ||
}} |
}} |
||
'''''Hepadnaviridae'''''{{efn|Etymology - portmanteau of ''hepa'' (liver: reference to Hepatitis B the primary human member) [[DNA]] virus.}} is a family of [[viruses]]. Humans, apes, and birds serve as natural hosts. It is a family of viruses that has the smallest genome among animal DNA viruses.<ref>{{Cite web|title=Hepadna Viral Family|url=https://web.stanford.edu/group/virus/hepadna/index.html|website=web.stanford.edu|access-date=2020-05-03}}</ref> There are currently 18 species in this family, divided among 5 genera. Its best-known member is [[hepatitis B virus]]. Diseases associated with this family include: [[liver]] infections, such as hepatitis, hepatocellular carcinomas (chronic infections), and cirrhosis.<ref name=ViralZone>{{cite web|title=Viral Zone|url=http://viralzone.expasy.org/all_by_species/9.html|publisher=ExPASy|access-date=15 June 2015}}</ref><ref name=ICTV>{{cite web|last1=ICTV|title=Virus Taxonomy: 2014 Release|url=http://ictvonline.org/virusTaxonomy.asp|access-date=15 June 2015}}</ref> Hepadnaviruses have some similar features with the retrovirus family: a) Hepatitis B virus uses the reverse transcriptase enzyme to convert |
'''''Hepadnaviridae'''''{{efn|Etymology - portmanteau of ''hepa'' (liver: reference to Hepatitis B the primary human member) [[DNA]] virus.}} is a family of [[viruses]]. Humans, apes, and birds serve as natural hosts. It is a family of viruses that has the smallest genome among animal DNA viruses.<ref>{{Cite web|title=Hepadna Viral Family|url=https://web.stanford.edu/group/virus/hepadna/index.html|website=web.stanford.edu|access-date=2020-05-03}}</ref> There are currently 18 species in this family, divided among 5 genera. Its best-known member is [[hepatitis B virus]]. Diseases associated with this family include: [[liver]] infections, such as hepatitis, hepatocellular carcinomas (chronic infections), and cirrhosis.<ref name=ViralZone>{{cite web|title=Viral Zone|url=http://viralzone.expasy.org/all_by_species/9.html|publisher=ExPASy|access-date=15 June 2015}}</ref><ref name=ICTV>{{cite web|last1=ICTV|title=Virus Taxonomy: 2014 Release|url=http://ictvonline.org/virusTaxonomy.asp|access-date=15 June 2015}}</ref> Hepadnaviruses have some similar features with the retrovirus family: a) Hepatitis B virus uses the reverse transcriptase enzyme to convert RNA into DNA, just like retroviruses. b) The function of gag, pol and env genes in retroviruses is the same as C, P and S genes in Hepadnaviruses. c) Both are enveloped viruses.<ref name="Hepadna Viral Family">{{Cite web|title=Hepadna Viral Family|url=https://web.stanford.edu/group/virus/hepadna/index.html|website=web.stanford.edu|access-date=2020-05-04}}</ref> |
||
==Taxonomy== |
==Taxonomy== |
||
Revision as of 14:49, 21 May 2020
| Hepadnaviridae | |
|---|---|

| |
| TEM micrograph showing Hepatitis B virus virions | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Pararnavirae |
| Phylum: | Artverviricota |
| Class: | Revtraviricetes |
| Order: | Blubervirales |
| Family: | Hepadnaviridae |
| Genera[1] | |
Hepadnaviridae[a] is a family of viruses. Humans, apes, and birds serve as natural hosts. It is a family of viruses that has the smallest genome among animal DNA viruses.[2] There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas (chronic infections), and cirrhosis.[3][4] Hepadnaviruses have some similar features with the retrovirus family: a) Hepatitis B virus uses the reverse transcriptase enzyme to convert RNA into DNA, just like retroviruses. b) The function of gag, pol and env genes in retroviruses is the same as C, P and S genes in Hepadnaviruses. c) Both are enveloped viruses.[5]
Taxonomy
Hepadnavirus family consists of 5 types of viruses: One is the human virus that infects the human and the other is the animal virus. Below are the names of virus types:[5]
Additionally, the African cichlid hepadnavirus (ACHBV) is described but not recognized as a species.[6]
History and discovery
Although liver diseases transmissible among human populations were identified early in the history of medicine, the first known hepatitis with a viral etiological agent was Hepatitis A, in the picornaviridae family. Hepatitis B Virus (HBV) was identified as an infection distinct from Hepatitis A through its contamination of measles, mumps, and yellow fever vaccines in the 1930s and 1940s. These vaccines contained HBV-infected human serum as a stabilizing agent. HBV was identified as a new DNA virus in the 1960s, followed a couple of decades later by the discovery of the flavivirus hepatitis C. HBV was first identified in the lab as the "Australia agent" by Blumberg and colleagues in the blood of an Aboriginal transfusion patient. This work earned Blumberg the 1976 Nobel Prize in Medicine.
Genome
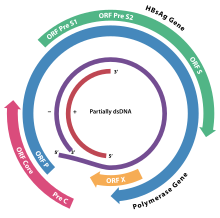
Genomes of hepadnaviruses consist of two DNA sequences, one of which is missing. Therefore, it is called partially single stranded and partially double stranded.[7] There is a complementary array at the 5 'ends. In this way, a short string has three strands. This provides a circular conformation to the DNA [7].Viral DNA chains are circular, but not closed.[8] The genome is very small, about 3.2 kb long.[7]
In the virion these strands are arranged such that the two ends of the long strand meet but are not covalently bonded together. The genome consists of two strands, a longer negative-sense strand and a shorter and positive-sense strand of variable length. The shorter strand overlaps this divide and is connected to the longer strand on either side of the split through a direct repeat (DR) segment that pairs the two strands together. In replication, the viral pdsDNA is converted in the host cell nucleus to covalently-closed-circular DNA (cccDNA) by the viral polymerase.
As it is a group 7 virus, replication involves an RNA intermediate. Four main open reading frames are encoded (ORFs) and the virus has four known genes which encode seven proteins: the core capsid protein, the viral polymerase, surface antigens—preS1, preS2, and S, the X protein (small X protein capable of transactivation of the transcription mechanism[9]). and HBeAg. The X protein is thought to be non-structural.
Viral polymerase
Hepadnaviridae encode their own polymerase, rather than co-opting host machinery as some other viruses do. This enzyme is unique among viral polymerases in that it has reverse transcriptase activity to convert RNA into DNA to replicate the genome (the only other human-pathogenic virus family encoding a polymerase with this capability is Retroviridae), RNAse activity (used when the DNA genome is synthesized from pgRNA that was packaged in virions for replication to destroy the RNA template and produce the pdsDNA genome), and DNA-dependent-DNA-polymerase activity (used to create cccDNA from pdsDNA in the first step of the replication cycle). Viral polymerase consummate the positive chain of rcDNA. Polymerase, located at the 5th end of the negative chain, is used for the positive chain. Short RNA-primer is used for positive DNA chain synthesis. Both the polymerase and short RNA-primer are removed via the cellular enzymes of the host. After the positive chain is completed, it mates with that negative chain and is covalently attached at both ends and creates a circular supercoil molecule [10]
Envelope proteins
The hepatitis envelope proteins are composed of subunits made from the viral preS1, preS2, and S genes. The L (for "large") envelope protein contains all three subunits. The M (for "medium") protein contains only preS2 and S. The S (for "small") protein contains only S. The genome portions encoding these envelope protein subuntis share both the same frame and the same stop codon (generating nested transcripts on a single open reading frame. The pre-S1 is encoded first (closest to the 5' end), followed directly by the pre-S2 and the S. When a transcript is made from the beginning of the pre-S1 region, all three genes are included in the transcript and the L protein is produced. When the transcript starts after the pro-S1 at the beginning of the pre-S2 the final protein contains the pre-S2 and S subunits only and therefore is an M protein. The smallest envelope protein containing just the S subunit is made most because it is encoded closest to the 3' end and comes from the shortest transcript. These envelope proteins can assemble independently of the viral capsid and genome into non-infectious virus-like particles that give the virus a pleomorphic appearance. These non-infectious particles are abundant in the blood of an infected person. The typical concentration of virion is 0.1 µg/ml and that amount for non-infectious particles (sphere like) is 100 µg/ml. This 1000x difference is not understood but it might be interpreted as these non-infectious particles are attracting the antibodies that produced as host cell's response. This action may be providing protection to virions from the host's immune response.[7]
Replication
Hepadnaviruses replicate through an RNA intermediate (which they transcribe back into cDNA using reverse transcriptase). The reverse transcriptase becomes covalently linked to a short 3- or 4-nucleotide primer.[11] Most hepadnaviruses will only replicate in specific hosts, and this makes experiments using in vitro methods very difficult.
The virus binds to specific receptors on cells and the core particle enters the cell cytoplasm. This is then translocated to the nucleus, where the partially double stranded DNA is 'repaired' by the viral polymerase to form a complete circular dsDNA genome (called covalently-closed-circular DNA or cccDNA). The genome then undergoes transcription by the host cell RNA polymerase and the pregenomicRNA (pgRNA) is sent out of the nucleus. The pgRNA is inserted into an assembled viral capsid containing the viral polymerase. Inside this capsid the genome is converted from RNA to pdsDNA through activity of the polymerase as an RNA-dependent-DNA-polymerase and subsequently as an RNAse to eliminate the pgRNA transcript. Small, medium and large envelope proteins locate inside of the endoplasmic reticulum during their translation and these proteins aggregate at the specific sites of the ER membrane of the host cell.[12] The assembled core migrates to the Golgi apparatus and interact with the Golgi membrane the site of enriched with surface proteins. It is said that LS protein interacts with the nucleocapsid and that enables the entry of the nucleocapsid inside the lumen of the Golgi. Later on the enveloped virions are released to extracellular environment via exocytosis.[13] These new virions either leave the cell to infect others or are immediately dismantled so the new viral genomes can enter the nucleus and magnify the infection. The virions that leave the cell egress through budding.
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Avihepadnavirus | Birds | Hepatocytes | Cell receptor endocytosis | Budding | Nucleus | Cytoplasm | Vertical: parental; sex; blood |
| Orthohepadnavirus | Humans; mammals | Hepatocytes | Cell receptor endocytosis | Budding | Nucleus | Cytoplasm | Vertical: parental; sex; blood |
Formation of the HBV capsid is coupled to reverse transcription. The capsid isn't merely AN inert “shell” as mutations in C have an effect on activities as various as DNA synthesis, transport of capsids back to the nucleus, and the surface of HBV glyproteins are encoded by the S ORF. The 3 cocarboxyl terminal molecules are created from 2 mRNAs and by use of different begin codons. Assembly of infectious particles needs interactions between the capsid and huge S within the cytoplasm. Intramission appears to occur by budding into the ER, with accumulation of virions in mutlivesicular bodies (MVB) of the late endosomal compartment. Particle unharness would then occur upon fusion of the MVB with the semipermeable membrane.[14]
Structure
Viruses in Hepadnaviridae are enveloped, with spherical geometries, and T=4 symmetry. The diameter is around 42 nm. Genomes are circular, around 3.2kb in length. The genome codes for 7 proteins.[3] Hepatitis DNA viruses (hepadnavirus) do not produce obvious surface outcomes after negative staining. Some hepadnaviruses are not spherical but may be plemorphic in nature.[15] They are enveloped and contain three large glycoproteins in their envelopes. The envelope covers the T = 4 icosahedral capsid. These capsids also have a diameter of approximately 34 nm. The DNA genome, partially double-wrapped and partially single-wrapped, is in the nucleus.[16] Replication in hepadnaviruses occurs by the synthesis of genomes by reverse transcription. The reverse transcription process is associated with capsid formation and this process takes place in the cytoplasm. Since the capsids have size limits, the genomes formed here are estimated to be deficient.[17] The whole thread in the genome is negatively between 3.0 and 3.3 kb in size. The size of the second strand is between 1.7 and 2.8 kb, and 15% of the genome is single stranded.[18]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Avihepadnavirus | Icosahedral | T=4 | Non-enveloped | Circular | Monopartite |
| Orthohepadnavirus | Icosahedral | T=4 | Non-enveloped | Circular | Monopartite |
Evolution
Based on the presence of viral genomes in bird DNA it appears that the hepadnaviruses evolved >82 million years ago.[19] Birds may be the original hosts of the Hepadnaviridae with mammals becoming infected after a bird (see host switch).
Endogenous hepatitis B virus genomes have been described in crocodilian, snake and turtle genomes.[20] This suggests that these viruses have infected vertebrates for over 200 million years ago.
Hepadnaviruses have been described in fish and amphibians also.[6] This suggests that this family has co-evolved with the vertebrates.
Phylogenetic trees suggest that the bird viruses originated from those infecting reptiles. Those affecting mammals appear to be more closely related to those found in fish.[21]
A family of viruses - the Nackednaviridae - has been isolated from fish.[21] This family has a similar genomic organisation to that of the Hepadnaviridae. These two families separated over 400 million years ago suggesting an ancient origin for the Hepadnaviridae.
Cell tropism
Hepadnaviruses, as their "hepa" name implies, infect liver cells and cause hepatitis. This is true not only of the human pathogen Hepatitis B Virus but also the hepadnaviruses that infect other organisms. The "adhesion" step of the dynamic phase—in which an exterior viral protein stably interacts with a host cell protein—determines cell tropism. In the case of HBV the host receptor is human sodium taurocholate receptor (NTCP), a mediator of bile acid uptake, and the virus anti-receptor is the abundant HB-AgS envelope protein.[22]
Notes
References
- ^ "Virus Taxonomy: 2018b Release". International Committee on Taxonomy of Viruses (ICTV). February 2019. Retrieved 14 March 2019.
- ^ "Hepadna Viral Family". web.stanford.edu. Retrieved 3 May 2020.
- ^ a b "Viral Zone". ExPASy. Retrieved 15 June 2015.
- ^ ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- ^ a b "Hepadna Viral Family". web.stanford.edu. Retrieved 4 May 2020.
- ^ a b Dill JA, Camus AC, Leary JH, Di Giallonardo F, Holmes EC, Ng TF (September 2016). "Distinct Viral Lineages from Fish and Amphibians Reveal the Complex Evolutionary History of Hepadnaviruses". Journal of Virology. 90 (17): 7920–33. doi:10.1128/JVI.00832-16. PMC 4988138. PMID 27334580.
- ^ a b c d Carter, John B., 1944- (2007). Virology : principles and applications. Saunders, Venetia A., 1949-. Chichester, England: John Wiley & Sons. ISBN 978-0-470-02386-0. OCLC 124160564.
{{cite book}}: CS1 maint: multiple names: authors list (link) CS1 maint: numeric names: authors list (link) - ^ Robinson, M.D, William S. (February 1994). "Molecular Events in the Pathogenesis of Hepadnavirus-Associated Hepatocellular Carcinoma". Annual Review of Medicine. 45 (1): 297–323. doi:10.1146/annurev.med.45.1.297. ISSN 0066-4219. PMID 8198385.
- ^ Robinson, M.D, William S. (February 1994). "Molecular Events in the Pathogenesis of Hepadnavirus-Associated Hepatocellular Carcinoma". Annual Review of Medicine. 45 (1): 297–323. doi:10.1146/annurev.med.45.1.297. ISSN 0066-4219. PMID 8198385.
- ^ İnan, Neşe, and Fehmi Tabak. "Hepatitis B Virus: Biology And Life Cycle". Viral Hepatit Dergisi, vol 21, no. 1, 2015, pp. 1-7. Galenos Yayinevi, doi:10.4274/vhd.36036.
- ^ Shin MK, Lee J, Ryu WS (June 2004). "A novel cis-acting element facilitates minus-strand DNA synthesis during reverse transcription of the hepatitis B virus genome". Journal of Virology. 78 (12): 6252–62. doi:10.1128/JVI.78.12.6252-6262.2004. PMC 416504. PMID 15163718.
- ^ Selzer, Lisa; Zlotnick, Adam (9 November 2015). "Assembly and Release of Hepatitis B Virus". Cold Spring Harbor Perspectives in Medicine. 5 (12): a021394. doi:10.1101/cshperspect.a021394. ISSN 2157-1422. PMC 4665036. PMID 26552701.
- ^ Acheson, N. H. (2011). Fundamentals of molecular virology (2nd ed.). Hoboken, NJ: John Wiley & Sons. ISBN 978-0-470-90059-8. OCLC 697768676.
- ^ Payne, S. (2017). Family Hepadnaviridae. Viruses, 321–327. doi:10.1016/b978-0-12-803109-4.00038-6
- ^ Hepadnaviridae. (2012). In Virus Taxonomy (pp. 445–455). Elsevier. https://doi.org/10.1016/b978-0-12-384684-6.00041-0
- ^ Other Viruses. (2017). In Fenner’s Veterinary Virology (pp. 547–555). Elsevier. https://doi.org/10.1016/b978-0-12-800946-8.00030-1
- ^ Payne, S. (2017). Family Hepadnaviridae. In Viruses (pp. 321–327). Elsevier. https://doi.org/10.1016/b978-0-12-803109-4.00038-6
- ^ Other Viruses. (2017). In Fenner’s Veterinary Virology (pp. 547–555). Elsevier. https://doi.org/10.1016/b978-0-12-800946-8.00030-1
- ^ Suh A, Brosius J, Schmitz J, Kriegs JO (2013). "The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses". Nature Communications. 4: 1791. Bibcode:2013NatCo...4.1791S. doi:10.1038/ncomms2798. PMID 23653203.
- ^ Suh A, Weber CC, Kehlmaier C, Braun EL, Green RE, Fritz U, Ray DA, Ellegren H (December 2014). "Early Mesozoic coexistence of amniotes and hepadnaviridae". PLOS Genetics. 10 (12): e1004559. doi:10.1371/journal.pgen.1004559. PMC 4263362. PMID 25501991.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Lauber C, Seitz S, Mattei S, Suh A, Beck J, Herstein J, Börold J, Salzburger W, Kaderali L, Briggs JA, Bartenschlager R (September 2017). "Deciphering the Origin and Evolution of Hepatitis B Viruses by Means of a Family of Non-enveloped Fish Viruses". Cell Host & Microbe. 22 (3): 387–399.e6. doi:10.1016/j.chom.2017.07.019. PMC 5604429. PMID 28867387.
- ^ Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W (November 2012). "Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus". eLife. 1: e00049. doi:10.7554/eLife.00049. PMC 3485615. PMID 23150796.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
External links
- Viralzone: Hepadnaviridae
- ICTV
- "Hepadnaviridae". NCBI Taxonomy Browser. 10404.

