Draft:Original research/Genomics

Genomics is a branch of molecular biology concerned with the structure, function, evolution, and mapping of genomes.
The H (heavy, outer circle) and L (light, inner circle) strands are given with their corresponding genes. There are 22 transfer RNA (TRN) genes for the following amino acids: F, V, L1 (codon UUA/G), I, Q, M, W, A, N, C, Y, S1 (UCN), D, K, G, R, H, S2 (AGC/U), L2 (CUN), E, T and P (white boxes). There are 2 ribosomal RNA (RRN) genes: S (small subunit, or 12S) and L (large subunit, or 16S) (blue boxes). There are 13 protein-coding genes: 7 for NADH dehydrogenase subunits (ND, yellow boxes), 3 for cytochrome c oxidase subunits (COX, orange boxes), 2 for ATPase subunits (ATP, red boxes), and one for cytochrome b (CYTB, coral box). Two gene overlaps are indicated (ATP8-ATP6, and ND4L-ND4, black boxes).
The control region (CR) is the longest non-coding sequence (grey box). Its three hyper-variable regions are indicated (HV, green boxes).
Genomes
[edit | edit source]
The genome is the entirety of an organism's hereditary information. In humans, it is encoded in DNA. The genome includes both the genes and the non-coding sequences of the DNA.[1]
Def. the "complete genetic information ... of an organism"[2] is called a genome.
Genetic information is encoded as a sequence of nucleobases: adenine (A), cytosine (C), guanine (G), and thymine (T).
There are "3.2 billion base pairs in the human genome."[3]
Associated with genomes are epigenomes.
Theoretical genomics
[edit | edit source]
Def. the "study of the complete genome of an organism"[4] is called genomics.
Sequencing
[edit | edit source]Genomics involves the sequencing and analysis of genomes through uses of high throughput DNA sequencing and bioinformatics to assemble and analyze the function and structure of entire genomes.[5][6][7]
Epistasis
[edit | edit source]Def. the "modification of the expression of a gene by another unrelated one"[8] is called epistasis.
Heterosis
[edit | edit source]Def. the "tendency of cross-breeding to produce an animal or plant with a greater hardiness than its parents; hybrid vigour"[9] is called heterosis.
Pleiotropy
[edit | edit source]Def. the "influence of a single gene on multiple phenotypic traits; pleiotropism"[10] is called pleiotropy.
Genomic diseases
[edit | edit source]"Genomics includes the scientific study of complex diseases such as heart disease, asthma, diabetes, and cancer because these diseases are typically caused more by a combination of genetic and environmental factors than by individual genes."[11]
Mitachondrial diseases
[edit | edit source]Genomic "mutations that cause mitochondrial disease [in T-cell mitochondria such as those imaged on the right] may also compromise affected people's immune response."[12]
Structural genomics
[edit | edit source]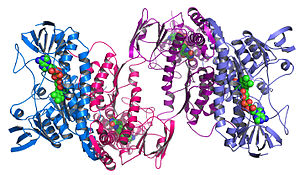
Structural genomics seeks to describe the 3-dimensional structure of every protein encoded by a given genome.[13][14]
This raises new challenges in structural bioinformatics, i.e. determining protein function from its 3D structure.[14]
Epigenomics
[edit | edit source]Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome.[15] Epigenetic modifications are reversible modifications on a cell’s DNA or histones that affect gene expression without altering the DNA sequence (Russell 2010 p. 475). Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis.[15] The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.[16]
Metagenomics
[edit | edit source]
Metagenomics is the study of metagenomes, genetic material recovered directly from environmental samples. The vast majority of microbial biodiversity had been missed by cultivation-based methods.[17] Recent studies use "shotgun" chain termination methods (Sanger sequencing) or massively parallel (pyrosequencing) to get largely unbiased samples of all genes from all the members of the sampled communities.[18] Because of its power to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world.[19][20]
As diagrammed in the image on the right, Environmental Shotgun Sequencing (ESS) is a key technique in metagenomics: (A) Sampling from habitat; (B) filtering particles, typically by size; (C) Lysis and DNA extraction; (D) cloning and library construction; (E) sequencing the clones; (F) sequence assembly into contigs and scaffolds.
Hypotheses
[edit | edit source]- The human genome has less than 40,000 isoforms.
See also
[edit | edit source]- A1BG gene transcriptions (127 kB) (30 September 2018)
- Agriculture (Middle Bronze Age) (15 kB) (13 September 2019)
- Ammonoids (62 kB) (22 September 2019)
- Animal physiology (8 kB) (12 September 2019)
- Anthropology (Aftonian, 540 ka) (30 kB) (29 March 2019)
- Biology (22 kB) (8 August 2019)
- Botany (82 kB) (24 February 2019)
- Culture (Aftonian) (9 kB) (28 October 2019)
- Cytogenetics (12 kB) (28 March 2019)
- Cytokinesis (12 kB) (29 October 2019)
- Dates (Hadean) (173 kB) (31 August 2019)
- Deoxyribonucleic acids (36 kB) (18 June 2019)
- Dominant group/Metagenome (65 kB) (28 March 2019)
- Epigenetics (27 kB) (28 May 2019)
- Epigenomes (27 kB) (28 May 2019)
- Eukaryotes (9 kB) (28 May 2019)
- Evolution (39 kB) (8 August 2019)
- Fruit and its importance (1 kB) (18 April 2019)
- Gene expressions in human exploration beyond low earth orbits (53 kB) (15 January 2019)
- Gene expressions/Project narrative (13 kB) (15 April 2018)
- Gene project (4 kB) (1 April 2019)
- Genes (23 kB) (4 November 2019)
- Genes/Expressions (3 kB) (29 March 2019)
- Hair colors (12 kB) (30 November 2018)
- Genetics (20 kB) (28 May 2019)
- Gene transcriptions (32 kB) (26 May 2019)
- Genomes (25 kB) (28 May 2019)
- Genomics (13 kB) (28 May 2019)
- Heredity (11 kB) (2 July 2019)
- Hominins (Miocene) (37 kB) (20 September 2019)
- Horticulture (3 kB) (24 July 2018)
- Human DNA (Paleogene) (168 kB) (19 August 2019)
- Human RNA (31 kB) (28 May 2019)
- Human skin pigmentation (4 kB) (16 August 2010)
- Human teeth (None) (30 kB) (2 July 2019)
- Lamarckism (20 kB) (25 July 2019)
- Libyan history (Oligocene) (29 kB) (13 August 2019)
- Mammalogy (Oligocene) (26 kB) (10 September 2019)
- Melanocytes (50 kB) (17 September 2019)
- Middle Ages (-10th century) (108 kB) (29 March 2019)
- Molecular genetics (11 kB) (28 May 2019)
- Osteoarthritis (12 kB) (31 August 2019)
- Paleanthropology (Paleogene) (195 kB) (30 August 2019)
- Paleontology (115 kB) (12 September 2019)
- Phosphate biochemistry (59 kB) (26 May 2019)
- Phosphate budgets (47 kB) (26 May 2019)
- Phosphate reactions (86 kB) (1 September 2019)
- Proteins (26 kB) (1 September 2019)
- Spaceflights (23 kB) (23 June 2019)
- Teeth (25 kB) (9 October 2019)
- What is a human? (23 kB) (10 September 2019)
- Zoology (178 kB) (1 October 2019)
References
[edit | edit source]- ↑ Ridley, M. (2006). Genome. New York, NY: Harper Perennial. ISBN 0-06-019497-9
- ↑ Msh210 (20 March 2009). genome. San Francisco, California: Wikimedia Foundation, Inc. http://en.wiktionary.org/wiki/genome. Retrieved 2012-10-30.
- ↑ Heidi Chial (2008). "DNA sequencing technologies key to the Human Genome Project". Nature Education 1 (1): 219. https://www.nature.com/scitable/nated/article?action=showContentInPopup&contentPK=828. Retrieved 2017-06-20.
- ↑ SemperBlotto (28 April 2007). genomics. San Francisco, California: Wikimedia Foundation, Inc. https://en.wiktionary.org/wiki/genomics. Retrieved 27 June 2014.
- ↑ National Human Genome Research Institute (8 November 2010). A Brief Guide to Genomics. http://www.genome.gov/19016904. Retrieved 2011-12-03.
- ↑ Concepts of genetics (10th ed.). San Francisco: Pearson Education. 2012. ISBN 978-0-321-72412-0.
- ↑ Richard Robinson, ed (8 November 2002). Genomics. Macmillan Reference USA. ISBN 978-0-02-865606-9.
- ↑ SemperBlotto (31 March 2007). epistasis. San Francisco, California: Wikimedia Foundation, Inc. https://en.wiktionary.org/wiki/epistasis. Retrieved 11 June 2017.
- ↑ SemperBlotto (17 March 2005). heterosis. San Francisco, California: Wikimedia Foundation, Inc. https://en.wiktionary.org/wiki/heterosis. Retrieved 11 June 2017.
- ↑ SemperBlotto (7 April 2008). pleiotropy. San Francisco, California: Wikimedia Foundation, Inc. https://en.wiktionary.org/wiki/pleiotropy. Retrieved 11 June 2017.
- ↑ Eric Green (2 March 2016). Frequently Asked Questions About Genetic and Genomic Science. Bethesda, Maryland USA: National Human Genome Research Institute. https://www.genome.gov/19016904/. Retrieved 11 June 2017.
- ↑ Peter McGuire (8 June 2017). "NHGRI study highlights role of mitochondria in immune response". Bethesda, Maryland USA: National Human Genome Research Institute. Retrieved 11 June 2017.
- ↑ "Towards a comprehensive structural coverage of completed genomes: a structural genomics viewpoint". BMC Bioinformatics 8: 86. March 2007. doi:10.1186/1471-2105-8-86. PMID 17349043. PMC 1829165. //www.ncbi.nlm.nih.gov/pmc/articles/PMC1829165/.
- ↑ 14.0 14.1 "Expectations from structural genomics". Protein Science 9 (1): 197–200. January 2000. doi:10.1110/ps.9.1.197. PMID 10739263. PMC 2144435. //www.ncbi.nlm.nih.gov/pmc/articles/PMC2144435/.
- ↑ 15.0 15.1 Francis, Richard C (2011). Epigenetics : the ultimate mystery of inheritance. New York: W.W. Norton. ISBN 978-0-393-07005-7.
- ↑ Laird PW (March 2010). "Principles and challenges of genomewide DNA methylation analysis". Nature Reviews. Genetics 11 (3): 191–203. doi:10.1038/nrg2732. PMID 20125086.
- ↑ Hugenholtz P, Goebel BM, Pace NR (September 1998). "Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity". Journal of Bacteriology 180 (18): 4765–74. PMID 9733676. PMC 107498. //www.ncbi.nlm.nih.gov/pmc/articles/PMC107498/.
- ↑ Eisen JA (March 2007). "Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes". PLoS Biology 5 (3): e82. doi:10.1371/journal.pbio.0050082. PMID 17355177. PMC 1821061. //www.ncbi.nlm.nih.gov/pmc/articles/PMC1821061/.
- ↑ Marco D, ed (2010). Metagenomics: Theory, Methods and Applications. Caister Academic Press. ISBN 978-1-904455-54-7.
- ↑ Marco D, ed (2011). Metagenomics: Current Innovations and Future Trends. Caister Academic Press. ISBN 978-1-904455-87-5.
External links
[edit | edit source]- African Journals Online
- Bing Advanced search
- GenomeNet KEGG database
- Google Books
- Google scholar Advanced Scholar Search
- Home - Gene - NCBI
- JSTOR
- Lycos search
- NCBI All Databases Search
- NCBI Site Search
- PubChem Public Chemical Database
- Questia - The Online Library of Books and Journals
- SAGE journals online
- Scirus for scientific information only advanced search
- SpringerLink
- Taylor & Francis Online
- WikiDoc The Living Textbook of Medicine
- Wiley Online Library Advanced Search
- Yahoo Advanced Web Search
|
Learn more about Genomics |
